Abstract
The x-ray structures of the unliganded esterase-like catalytic antibody D2.3 and its complexes with a substrate analogue and with one of the reaction products are analyzed. Together with the structure of the phosphonate transition state analogue hapten complex, these crystal structures provide a complete description of the reaction pathway. At alkaline pH, D2.3 acts by preferential stabilization of the negatively charged oxyanion intermediate of the reaction that results from hydroxide attack on the substrate. A tyrosine residue plays a crucial role in catalysis: it activates the ester substrate and, together with an asparagine, it stabilizes the oxyanion intermediate. A canal allows facile diffusion of water molecules to the reaction center that is deeply buried in the structure. Residues bordering this canal provide targets for mutagenesis to introduce a general base in the vicinity of the reaction center.
Keywords: phosphonate hapten, ester hydrolysis, oxyanion hole, catalytic antibody, catalysis
The proposal that antibodies having catalytic activity can be generated to an analogue of a transition state (TSA) of the reaction to be catalyzed (1) has proven to be widely applicable (2, 3). Much has been learned over the last several years about inducing antibodies that promote ester hydrolysis: more than 50 anti-phosphonate antibodies with esterolytic activity have been reported to date. Although extensive steady-state and pre-steady-state kinetic studies have been performed on some antibodies (see ref. 4), the available structural data have focused on the interactions of esterolytic antibodies with phosphonate TSAs (5–8). These confirm the exquisite shape and chemical complementarity between the binding pocket and the haptenic TSA; however, these studies shed little light on the exact pathway that leads substrate to product in an antibody-mediated reaction.
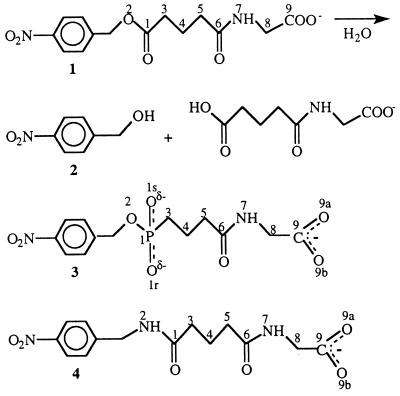
Antibody D2.3 catalyses the hydrolysis of the nonactivated p-nitrobenzyl ester 1 (Fig. 1); it was obtained by immunizing BALB/c mice with a protein conjugate of the phosphonate hapten 3 and identified by screening the entire repertoire of hybridomas for catalytic activity (9). D2.3 is the most efficient of the family of antibodies obtained and it accelerates the target reaction by a factor of 105 (10). We previously determined the x-ray structure of the Fab D2.3 complexed with TSA 3 at 1.9-Å resolution (8). We now report the crystal structures of the Fab D2.3 unliganded, complexed with p-nitrobenzyl amide 4 (a substrate analogue inhibitor, SA) and with one of the products of the reaction, p-nitrobenzyl alcohol 2; this allows us to describe key stages along the pathway of the catalyzed reaction. Together with the Fab–TSA 3 structure, the new data support a mechanism whereby hydroxide attacks the carbonyl of the scissile ester bond in the substrate, identify the groups that stabilize the oxyanion intermediate, and define a pathway for the water molecule that gives rise to the attacking hydroxide ion. Only antibody-based hapten-programmed catalysts, such as D2.3, can be considered for comparing the substrate–catalyst interactions and those between this catalyst and a structure to which tight binding has been elicited, namely the TSA.
Figure 1.
Chemical reaction catalyzed by D2.3 and structures of the compounds used in this study. Ester 1 is a substrate hydrolyzed by D2.3. Crystal structures examined are those of complexes of D2.3 with p-nitrobenzyl alcohol 2 (one of the products of the hydrolysis of 1), p-nitrobenzyl phosphonate 3 (the TSA hapten used to elicit D2.3), and p-nitrobenzyl amide 4, a stable analogue of the substrate ester 1.
MATERIALS AND METHODS
Purification, Crystallization, and Data Collection.
The D2.3 antibody was obtained and propagated in ascitic fluids as described (9). The preparation and the purification of D2.3 IgG was similar to described protocols (11) and resulted in fully active IgG, as judged by titration of catalytic activity with 3. After dialysis in Tris-buffered saline (pH 7.5), the IgG was concentrated to 2 mg/ml and proteolyzed with papain under standard conditions [1% (wt/wt) papain to IgG ratio; digestion time, 2.5 h] to produce the Fab. Undigested IgG and Fc fragment were removed by protein A chromatography, and the Fab was further purified by gel filtration on a Sephacryl S100 HR column.
For crystallization the Fab was concentrated to 5 mg/ml and complexed with ligand in 5- to 10-fold excess. Crystallizations were performed following the hanging drop method, and initial conditions were identified by sparse matrix sampled screening (12). To get better diffracting crystals, polyethylene glycol (PEG) 600 precipitant [≈30% (wt/vol)] was used together with 40 mM of Zn acetate. The protein concentration (5–10 mg/ml) and the pH (100 mM cacodylate buffer, pH 6.0–7.5) were less critical. The crystals (space group P3121) diffract at least to 2.0-Å resolution. Unliganded Fab crystallizes at higher PEG 600 concentration [35% (wt/vol)], and data were collected to 2.5 Å. Diffraction data were collected in each case from one crystal at the D41 and W32 stations of the Laboratoire pour l’Utilisation du Rayonnement Electromagnétique synchrotron (Orsay, France) using a Mar-Research (Hamburg, Germany) image plate. The data were indexed with denzo (13), integrated with mosflm (14), scaled, and merged with programs of the CCP4 suite (15). Statistics are reported in Table 1.
Table 1.
Crystallographic data and refinement statistics
| Unliganded D2.3 | D2.3–4 SA | D2.3–2 product | D2.3–3 TSA | |
|---|---|---|---|---|
| Data collection statistics | ||||
| Unit cell | ||||
| a (=b), c (Å) | 78.5, 158.9 | 78.2, 158.7 | 78.5, 159.5 | 78.2, 158.9 |
| α (=β), γ (°) | 90, 120 | 90, 120 | 90, 120 | 90, 120 |
| Resolution limit, Å | 2.55 | 2.0 | 2.0 | 1.9 |
| Observations | 57,600 | 89,942 | 98,164 | 92,749 |
| Unique reflections | 18,580 | 38,114 | 39,071 | 44,679 |
| Completeness* % | 96.0 (76.0) | 98.8 (98.7) | 99.6 (98.6) | 98 (97) |
| Rsym*† % | 4.8 (17) | 5.5 (30.1) | 5.2 (31.3) | 6.3 (33) |
| I/σ(I)* | 11.6 (4.4) | 7.8 (2.5) | 10.1 (2.4) | 5.6 (2.1) |
| Refinement statistics | ||||
| Resolution, Å | 7.0 − 2.55 | 7.0 − 2.0 | 7.0 − 2.0 | 7.0 − 1.9 |
| R‡ % | 18.1 | 19.9 | 19.9 | 20.8 |
| Rfree, % | 23.1 | 24.8 | 24.0 | 25.2 |
| Water molecules | 130 | 160 | 154 | 149 |
| rms deviations from ideality | ||||
| Bond lengths,§ Å | 0.008 | 0.01 | 0.009 | 0.009 |
| Bond angles, (°)§ | 1.6 | 1.7 | 1.65 | 1.7 |
| Ramachandran core,¶ % | 89.0 | 90.8 | 89.5 | 89.8 |
Number in parentheses is the value in the highest resolution shell.
Rsym = ΣΣj|Ij − 〈I〉|/Σ|〈I〉|, where Ij is the intensity measurement for reflection j and 〈I〉 is the mean intensity for multiply recorded reflections.
R = Σ∥Fobs| − |Fcalc∥/Σ|Fobs|.
Evaluated with x-plor (16).
Evaluated with procheck (17).
Structure Determination and Refinement.
All the crystal forms of complexed and unliganded Fab reported here are isomorphous. Crystallographic refinements of the new D2.3 structures were performed with the conjugate-gradient facility of x-plor (16). The starting point was a model of Fab D2.3 complexed with TSA 3 and refined to an factor R of 21.5% and a free factor R (Rfree) of 26% in the 7- to 1.9-Å resolution range (8); before refinement of the new structures, water molecules and the ligand were removed from the model. The progress of the refinement was judged by decrease of Rfree after Powell minimization and temperature factor refinement (18). After initial refinement, 2Fobs − Fcalc and Fobs − Fcalc electron density maps with calculated phases were examined using program o (19) and showed clear electron density for the p-nitrobenzyl part of the amide 4 and for the whole p-nitrobenzyl alcohol molecule 2.
Ligand and water molecules were included in subsequent refinement cycles. Solvent molecules were identified with program o at positions of peaks higher than 4.5 standard deviations in Fobs − Fcalc maps and within hydrogen bond distance of polar Fab atoms. At the end of the refinement, electron density is continuous for all ligands. The final R and Rfree factors are reported in Table 1; the structures have standard stereochemistry, as evaluated with procheck (17).
Superposition of atomic models was done with program o (19). Surface areas were calculated using the algorithm of Shrake and Rupley (20). The solvent cavities were investigated with the program voidoo (21).
RESULTS AND DISCUSSION
Overall Structure of the Combining Site.
In all of the three D2.3 complex structures, the ligand is located in a deep pocket in the antibody combining site at the interface between the heavy and light chain variable regions. In each complex at least ≈90% of the accessible surface of the ligand is buried. The bottom of the pocket, where the p-nitrobenzyl moiety of the ligand is found, has a marked hydrophobic character: seven residues make apolar contacts with the p-nitrobenzyl group. The tight Van der Waals interactions between p-nitrobenzyl and the Fab account for the specificity of D2.3 for para- relative to ortho-nitrosubstituted ligands (10), because the corresponding change in the position of substitution would require a significant rearrangement of the Fab residues to be accommodated.
Consistent with the linkage of hapten 3 to the carrier protein through its N-glutarylglycinate carboxylic acid group (9), the carboxylates of ligands 3 and 4 point toward the outside of the combining site. The overall orientation of the ligand relative to the antibody is similar to those observed in complexes of antibodies D2.4 and D2.5, which catalyze the same reaction as D2.3 (8), as well as in complexes of the three other esterolytic antibodies whose crystallographic structures have been determined (5–7). This confirms the suggestion made previously that this mode of binding is favored when the hapten comprises proximal aromatic and phosphonate moieties (6), although slight variations in the orientation of the aromatic groups have been noted (22).
The flexibility of the combining site can be discussed based on a superposition of the structures of the Fab D2.3 that have been determined (unliganded and liganded either with the amide 4, the phosphonate 3, or the alcohol 2). In all four structures, the residues of the combining site superimpose well within experimental error and no significant movement occurs upon ligand binding [rms deviation of the Cαs of the 65 residues of the complementarity determining regions (CDRs) after superposition: 0.2 Å or less]; this differs from what has been observed in other antibodies raised against small molecules or against proteins (23). The same conformation of the combining site, including that of its most flexible H3 CDR loop, is also found in the complex of antibody D2.5 with TSA 3; D2.5 was raised in the same fusion as D2.3 and crystallizes with a packing different from that of D2.3 (8) (the H3 loop of D2.5 has the same length and the same sequence as that of D2.3 except for the semi-conservative H102 Met → Leu mutation). This suggests that the similarity of the structures of the three complexes we studied does not reflect specific packing constraints, but is due to a stable conformation of the complexed combining site, and in particular of its H3 CDR.
Substrate Binding.
Amide 4 is a SA inhibitor of D2.3 (9), the difference between this compound and ester 1 being the replacement by an NH group of the oxygen atom of the ester bond that is hydrolyzed by D2.3. Amide and ester bonds differ in their hydrogen bonding requirements, but apart from that, on the basis of small molecule x-ray structures, the geometries of compounds 1 and 4 should be nearly identical. In the complex of 4 with D2.3, the N2H group (numbering as in Fig. 1) is not within hydrogen bonding distance with the Fab (distance to the closest atom, 3.6 Å) (Fig. 2A). The oxygen O2 of 1 could not establish a hydrogen bond with D2.3 without a significant rearrangement of the combining site because of the constraints associated with the positioning of the aromatic and glutarylglycinate moieties of the substrate. Given the close similarity of the four structures of D2.3 we determined, such a rearrangement is unlikely. Therefore, it is most likely that the ester oxygen that occupies the same position in 1 as the N2H in 4 would be found at the same location in the complex with D2.3 and the Fab–SA 4 complex may be considered a faithful mimic of the Fab–1 Michaelis–Menten complex.
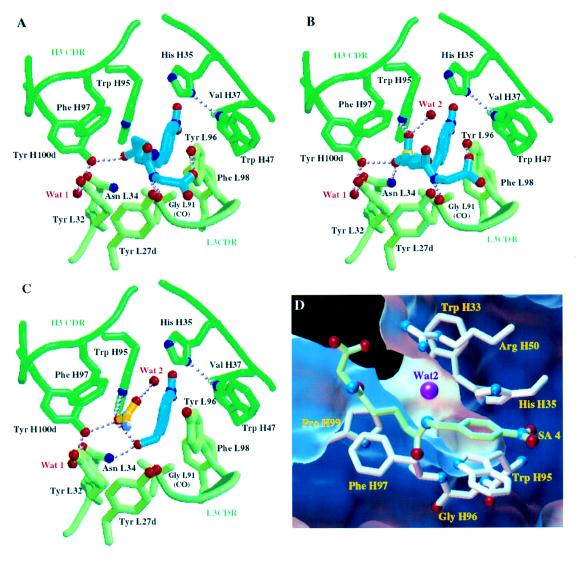
Figure 2.
Schematic views of D2.3 Fab residues that interact with the ligands examined. Residue numbering is according to ref. 24. In A–C, the ligands are in blue, the Cα trace of the Fab is in green, and water molecules are in red. Hydrogen bonds are shown as dotted lines. (A) Complex of D2.3 with amide 4, a stable SA. (B) Complex of D2.3 with TSA 3. (C) Complex of D2.3 with the reaction product 2, p-nitrobenzyl alcohol. Electron density corresponding to an acetate molecule was located in the combing site. The acetate is in yellow; the oxygens and the methyl of the acetate were distinguished on the basis of the hydrogen bonds established. A–C were drawn with molscript (25). (D) View of the canal (D2.3-4 structure) that allows water diffusion to the carbon atom of the carbonyl of 4 analogous to the reaction center in the complex of D2.3 with 1. The surface accessible to the exterior of a water molecule represented by a 1.4-Å radius sphere is cut to show the canal; its face toward the complexed Fab atoms is in blue, and the one facing the exterior is in white. Only the residues that border the canal are represented. Ligand 4 is in green; the water molecule (magenta) closest to the analogue of the reaction center is within hydrogen bonding distance and angle to Arg-H50 (hydrogen bond not shown). Nitrogen Nδ1 of His-H35 makes a hydrogen bond to Trp-H47 (not shown) that is conserved in antibodies; therefore, the Nɛ2 nitrogen of His-H35 (which is part of the canal’s wall) is protonated, and His-H35 most likely does not function as a general base in the hydrolysis catalyzed by D2.3. D was rendered in the avs environment (26).
The major part of SA 4 is buried in the complex (only 10% of its surface remains accessible), so that multiple Van der Waals interactions are made with the Fab, including those of the p-nitrobenzyl moiety that have been described above. In addition, several hydrogen bonds stabilize the SA. In particular, the carbonyl oxygen O1 (Fig. 1), equivalent to the carbonyl oxygen of the ester bond in substrate 1, is within hydrogen bond distance (2.65 Å) and angle to the hydroxyl of Tyr-H100d (Fig. 2A). Two other hydrogen bonds are made with atoms of the N-glutarylglycinate part of 4 (Fig. 1, atoms C3 to O9). One of them is between N7 of the SA and the carbonyl oxygen of Gly-L91, the other bond is between O9 of the SA and the hydroxyl of Tyr-L96.
The N-glutarylglycinate chain, both in the SA and in the TSA, has a less well-defined electron density than the rest of the ligand. This is reflected by an average B factor of this part of the SA which is 10 Å2 higher than that for the whole molecule. This suggests that the N-glutarylglycinate binds less efficiently to D2.3 than the p-nitrobenzyl moiety or the reaction center mimic—i.e., the phosphonate or amide groups. The resulting stabilization is nevertheless significant because p-nitrobenzyl acetate, a short substrate that cannot establish hydrogen bonds with Gly-L91 or Tyr-L96, was found to be hydrolyzed significantly less efficiently than 1 [kcat/KM estimated to be less than 10 M−1⋅s−1 for this short substrate (9), as compared with 185 M−1⋅s−1 for substrate 1 (10)]. Because the two hydrogen bonds that stabilize the N-glutarylglycinate part of SA 4 are also observed in the complex of Fab D2.3 with TSA 3 (Fig. 2B), they contribute both to substrate and to transition state binding.
Interactions of the Reaction Products with the Combining Site.
When associated with D2.3, the p-nitrobenzyl alcohol 2 superposes within experimental error with the corresponding part of the SA, except for its hydroxyl group which makes a hydrogen bond with Asn-L34 (Fig. 2 A and C). The interactions of p-nitrobenzyl alcohol with D2.3 Fab residues account for the noticeable product inhibition by 2 observed during hydrolysis of 1 [estimated Ki, 50 μM (9)]. Additional and unexpected interactions are made between the p-nitrobenzyl hydroxyl group and a solvent molecule whose electron density corresponds to that of an acetate ion present in the crystallization solution (at 40 mM concentration). The acetate ion was refined with full occupancy and its final average B factor is identical to that of p-nitrobenzyl alcohol (B = 27 Å2); this suggests that the p-nitrobenzyl alcohol/acetate stoichiometry in the crystallized complex is 1:1. The N-glutarylglycinate acid product can be modeled in the combining site by superposing the glutarate carboxylate with that of the acetate found in the structure while avoiding unfavorable contacts with the Fab. Therefore, the position of the carboxylate of this acetate might indicate the localization of the nascent acid resulting from hydrolysis of ester 1 catalyzed by D2.3.
Catalytic Mechanism.
Based on the structure of Fab–TSA 3, we proposed that D2.3 acts mainly by stabilizing the negatively charged intermediate formed after hydroxide attack on the ester (8). Catalytic antibodies are designed such as to better stabilize the transition state of a reaction than its substrate. Given the structures of complexes of D2.3 with SA 4 and TSA 3, we are in a position to analyze how such differential stabilization is achieved. There is no sizable conformational change of the Fab during the reaction (see above). The ligands are best compared by considering separately the p-nitrobenzyl part, the glycinate end, and the glutaryl moiety (in which we include the reaction center analogue, a phosphonate in 3 and an amide in 4). The p-nitrobenzyl parts of the ligands superpose within experimental error (rms deviation of 10 atoms, 0.25 Å) and the positions of the glycinate end, which is tethered by a hydrogen bond to Gly-L91 in both structures (Fig. 2 A and B), closely overlap, too. The noticeably distinct planar amide of 4 and tetrahedral phosphonate in 3 can be accommodated in between the two conserved ends of the ligands due to the conformational flexibility of the glutaryl moiety. The resulting positional difference of the glutaryl methylenes C3 (Fig. 1) adjacent to the phosphonate in 3 and to the glutaryl carbonyl in 4 is 1.95 Å; this is the most prominent positional change on the reaction path.
Whereas the negatively charged phosphonate oxygens in 3 establish three hydrogen bonds with the antibody (Fig. 2B), the corresponding neutral amide in 4 makes only one such interaction (Fig. 2A). The preferential stabilisation of the TSA as compared with SA (three hydrogen bonds involving charged atoms vs. one hydrogen bond established by neutral atoms) is consistent with the ratio of the binding constants of TSA and substrate [1.1 × 105, measured at pH 6 (10)].
In the Fab–SA 4 structure, Tyr-H100d makes a hydrogen bond with the oxygen O1 of the SA 4 equivalent to the carbonyl oxygen of the to-be-hydrolyzed ester bond in 1, thus orienting this oxygen for later stabilization of the hydrolysis oxyanion intermediate by Tyr-H100d and Asn-L34 and activating the ester carbonyl for subsequent attack by a hydroxide ion. This Tyr-H100d–O1 hydrogen bond differentiates the two faces of the ester carbonyl, blocking access of hydroxide ions to one of them. Furthermore, a funnel-shaped cavity beside the alkyl chain of the SA 4 allows hydroxide ions to diffuse toward the other face of this same carbonyl (Fig. 2D). A water molecule (Wat2, Fig. 2B) well defined (Bfact: 17 Å2) in the Fab–TSA 3 structure and within hydrogen bonding distance to the pro-R phosphonyl oxygen provides a picture of the hydroxide en route toward the ester carbonyl carbon atom. Taken together, the D2.3–TSA and D2.3–SA structures define the Tyr-H100d hydroxyl and the Asn-L34 amide as the oxyanion hole, as well as the path followed by the water molecule that gives rise to the attacking hydroxide ion. Thus they define the mechanism for catalysis by D2.3 of direct hydroxide attack on ester 1.
Improvement of Catalytic Efficiency.
The value of the catalytic acceleration of antibody D2.3 (kcat/kuncat = 1.1 × 105) (10) places this antibody among the most efficient of those with an esterase-like activity. The structures determined here allow us to identify two features that account at least in part for this efficiency: the antibody is poised to stabilize both the SA and the TSA so that no significant rearrangement of the protein seems to be required to go from substrate to transition state binding; and directed hydrogen bonds (established by residues Asn-L34 and Tyr-H100d, which belong to the L1 and H3 CDRs) stabilize the oxyanion intermediate more efficiently than the substrate. It is remarkable that D2.3 does not make use of cationic residues that have been used in other catalytic antibodies whose structure have been determined (5, 7); these are well known to stabilize oxyanions more efficiently than tyrosines (27), which are rarely used for that purpose. Replacement of Tyr-H100d by an arginine or a lysine would therefore be expected to improve activity; structural adjustments would undoubtedly be required to optimally position the positive charge of the mutated residue. It remains to be seen whether they would be tolerated by the combining site or, as is more likely, whether additional mutations would be needed to allow such adjustments.
The canal that allows water to diffuse to the reaction center defines additional targets for mutagenesis. In D2.3 none of the residues bordering this canal is able to facilitate deprotonation of incoming water molecules. Among these residues, Arg-H50, His-H35, and Trp-H33 (which belong to CDR2 and CDR1 of the heavy chain) make the canal’s wall facing the accessible face of the hydrolyzed ester carbonyl, where the attacking water is located (Fig. 2D); because their side chains contact the solvent, they are structurally less constrained than, for instance, residues of the oxyanion hole. These residues are logical candidates for site specific mutagenesis to provide a general base in the vicinity of the reaction center. When evolving D2.3 through mutations of the oxyanion hole or of residues bordering the water canal, one would want to shift the pressure from tight binding to 3 (which has been a determining factor during maturation of the immune response that gave rise to this antibody) to catalytic efficiency. Methods developed to diversify the CDRs (28) coupled to a screening for catalytic activity (9, 29) are well suited for that purpose.
Acknowledgments
We thank B. Golinelli, J. Janin, F. Lederer, and A. Ménez for reading the manuscript. We thank M. Pique for his enthusiastic help in making Fig. 2. We are grateful to A. Bentley and J. Perez for helping us to use facilities at the Laboratoire pour l’Utilisation du Rayonnement Electromagnétique (Orsay, France). This work was supported in part by Contract 94/128 from the Direction de la Recherche Et de la Technologie.
ABBREVIATIONS
- CDR
complementarity determining region
- SA
substrate analogue
- TSA
transition state analogue
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, Chemistry Department, Brookhaven National Laboratory, Upton, NY 11973 (references IYEC, IYEF, IYEG, and IYEH).
References
- 1.Jencks W P. Catalysis in Chemistry and Enzymology. New York: McGraw–Hill; 1969. [Google Scholar]
- 2.Lerner R A, Benkovic S J, Schultz P G. Science. 1991;252:659–667. doi: 10.1126/science.2024118. [DOI] [PubMed] [Google Scholar]
- 3.Thomas N R. Nat Prod Rep. 1996;13:479–511. doi: 10.1039/np9961300479. [DOI] [PubMed] [Google Scholar]
- 4.Benkovic S J, Adams J A, Borders C L, Janda K D, Lerner R A. Science. 1990;250:1135–1139. doi: 10.1126/science.2251500. [DOI] [PubMed] [Google Scholar]
- 5.Zhou G W, Guo J, Huang W, Fletterick R J, Scanlan T S. Science. 1994;265:1059–1064. doi: 10.1126/science.8066444. [DOI] [PubMed] [Google Scholar]
- 6.Charbonnier J-B, Carpenter E, Gigant B, Golinelli-Pimpaneau B, Eshhar Z, Green B S, Knossow M. Proc Natl Acad Sci USA. 1995;92:11721–11725. doi: 10.1073/pnas.92.25.11721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patten P A, Gray N S, Yang P L, Marks C B, Wedemayer G J, Boniface J J, Stevens R C, Schultz P G. Science. 1996;271:1086–1091. doi: 10.1126/science.271.5252.1086. [DOI] [PubMed] [Google Scholar]
- 8.Charbonnier J-B, Golinelli-Pimpaneau B, Gigant B, Tawfik D S, Chap R, Schindler D S, Kim S-H, Green B S, Eshhar Z, Knossow M. Science. 1997;275:1140–1142. doi: 10.1126/science.275.5303.1140. [DOI] [PubMed] [Google Scholar]
- 9.Tawfik D S, Green B S, Chap R, Sela M, Eshhar Z. Proc Natl Acad Sci USA. 1993;90:373–377. doi: 10.1073/pnas.90.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tawfik D S, Lindner A, Chap R, Eshhar Z, Green B S. Eur J Biochem. 1997;244:619–626. doi: 10.1111/j.1432-1033.1997.00619.x. [DOI] [PubMed] [Google Scholar]
- 11.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 12.Jancarik J, Kim S. J Appl Crystallogr. 1991;24:409–411. [Google Scholar]
- 13.Otwinowsky Z, Minor W. Methods Enzymol. 1997;276:307–325. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 14.Leslie, A. G. W. (1992) CCP4 ESF-EACMB Newsl. Protein Crystallogr. 26.
- 15.CCP4. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 16.Brünger A T. x-plor Manual. New Haven, CT: Yale Univ.; 1992. , Version 3.1. [Google Scholar]
- 17.Laskowski R A, McArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 18.Brünger A T. Nature (London) 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 19.Jones T A, Zhou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 20.Shrake A, Rupley J A. J Mol Biol. 1973;79:351–371. doi: 10.1016/0022-2836(73)90011-9. [DOI] [PubMed] [Google Scholar]
- 21.Kleywegt G J, Jones T A. Acta Crystallogr D. 1994;50:178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- 22.MacBeath G, Hilvert D. Chem Biol. 1996;3:433–445. doi: 10.1016/s1074-5521(96)90091-5. [DOI] [PubMed] [Google Scholar]
- 23.Stanfield R L, Takimoto-Kamimura M, Rini J M, Profy A T, Wilson I A. Structure. 1993;1:83–93. doi: 10.1016/0969-2126(93)90024-b. [DOI] [PubMed] [Google Scholar]
- 24.Kabat E. Sequences of Proteins of Immunological Interest. Washington, DC: National Institute of Health; 1991. [Google Scholar]
- 25.Kraulis P. J Appl Crystallogr. 1991;24:924–950. [Google Scholar]
- 26.Upson C. IEEE Comput Graph Appl. 1989;9:30. [Google Scholar]
- 27.Phillips M A, Fletterick R, Rutter W J. J Biol Chem. 1990;265:20692–20698. [PubMed] [Google Scholar]
- 28.Schier R, Mccall A, Adams G P, Marshall K W, Merritt H, Yim M, Crawford R S, Weiner L M, Marks C, Marks J D. J Mol Biol. 1996;263:551–567. doi: 10.1006/jmbi.1996.0598. [DOI] [PubMed] [Google Scholar]
- 29.Janda K D, Lo L C, Lo C H L, Sim M M, Wang R, Wong C H, Lerner R A. Science. 1997;275:945–948. doi: 10.1126/science.275.5302.945. [DOI] [PubMed] [Google Scholar]