Summary
Ca2+ influx through voltage-gated channels initiates the exocytotic fusion of synaptic vesicles to the plasma membrane. Here we show that RIM-binding proteins (RBPs), which associate with Ca2+ channels in hair cells, photoreceptors, and neurons, interact with α1D (L-type) and α1B (N-type) Ca2+-channel subunits. RBPs contain three Src homology 3 domains that bind to proline-rich motifs in α1 subunits and Rab3-interacting molecules (RIMs). Overexpression in PC12 cells of fusion proteins that suppress the interactions of RBPs with RIMs and α1 augments the exocytosis triggered by depolarization. RBPs may regulate the strength of synaptic transmission by creating a functional link between the synaptic-vesicle tethering apparatus, which includes RIMs and Rab3, and the fusion machinery, which includes Ca2+ channels and the SNARE complex.
Introduction
The secretion of neurotransmitters and hormones is triggered by Ca2+ influx through voltage-gated channels. Cytoplasmic Ca2+ triggers a fusion reaction between vesicles and the plasma membrane whose central component is the SNARE complex comprising the vesicle-associated protein VAMP/synaptobrevin and the plasma-membrane proteins syntaxin and SNAP-25. This core complex interacts with additional proteins that modulate SNARE assembly and dissociation (reviewed in Lin and Scheller, 2000; Mochida, 2000). The SNARE complex also interacts with the Ca2+-binding protein synaptotagmin, which initiates vesicle fusion, and with snapin, which regulates the SNARE-synaptotagmin interaction (reviewed in Geppert and Südhof, 1998; Mochida, 2000). Finally, SNARE proteins associate with α1B (CaV2.2) N-type and α1A (CaV2.1) P/Q-type subunits of voltage-gated Ca2+ channels in neurons and with α1C (CaV1.2) and α1D (CaV1.3) L-type subunits in endocrine cells (reviewed in Atlas, 2001; Catterall, 1999). By tethering the fusion machinery near the site of Ca2+ influx, these interactions speed and synchronize synaptic transmission and hormone release.
Vesicle release is modulated by the GTP-binding protein Rab3 and the associated proteins rabphilin (Shirataki et al., 1993), Noc2 (Haynes et al., 2001), and RIM1 (Wang et al., 1997). Rab3 is a negative regulator of exocytosis (reviewed in Geppert and Südhof, 1998). In its GTP-bound form, Rab3 is associated with synaptic vesicles as well as with rabphilin, Noc2, and RIM1. Because RIM1 is specifically associated with the synaptic plasma membrane at the active zone, it may act as a regulator of vesicle fusion by inducing the formation of a GTP-dependent complex between synaptic vesicles and the plasma membrane (Wang et al., 1997).
Hair cells and photoreceptors ordinarily do not produce action potentials, so their afferent synapses release transmitter in response to graded receptor potentials. Because at threshold these signals are smaller than 1 mV, the vesicle-release machinery must be especially sensitive. The afferent synapses of hair cells are also unusual in that those in the auditory system can transmit information at frequencies as great as 10 kHz, whereas those in the vestibular system can signal steady accelerations indefinitely. Photoreceptors, too, are required to release neurotransmiter continuously in darkness. Presumably to facilitate their particular functions, the afferent synapses of receptor cells are morphologically specialized. Each presynaptic active zone in a hair cell comprises a spherical or ovoid dense body, surrounded by a halo of lucent synaptic vesicles, and apposed to a prominent presynaptic membrane density (Hama, 1980). In a photoreceptor, vesicles are instead packed against a broad, flattened synaptic ribbon (Zenisek et al., 2000).
Although the biochemical features of synapses in sensory receptors must be adapted for the cells’ specialized roles, little is known about the molecular constituents of the presynaptic active zones in these cells. In both hair cells and photoreceptors, the Ca2+ current that triggers exocytosis is borne by L-type Ca2+ channels (Corey et al., 1984; Roberts et al., 1990; Zidanic and Fuchs, 1995), rather than the N- and P/Q-type channels that predominate in synapses of the central nervous system. The hair cell’s L-type Ca2+ channels, which contain unique domains owing to an unusual pattern of mRNA splicing (Kollmar et al., 1997a, b; Platzer et al., 2000), have a relatively negative threshold of activation, rapid activation and deactivation kinetics, and little or no Ca2+-dependent inactivation (Hudspeth and Lewis, 1988; Zidanic and Fuchs, 1995). To seek other novel constituents of sensory synapses, we have searched for proteins that interact with the modified α1D Ca2+-channel subunit in hair cells. The protein identified in this endeavor appears to be involved in vesicle release, not only at the synapses of hair cells and photoreceptors, but also at those of many neurons.
Results
Identification of RBP2 as an α1D-binding protein
To identify proteins that bind to the α1D subunit of L-type Ca2+ channels in the inner ear, we performed a yeast two-hybrid screen with the cytoplasmic carboxyl-terminal domain of α1D as bait. Screening of a cDNA library derived from the sensory epithelium of the chicken’s cochlea, the basilar papilla, led to the isolation of seven clones of similar sequence (Fig. 1A). These clones closely resembled the human expressed-sequence tag KIAA0318 and a rat cDNA that encodes the carboxyl terminus of a protein termed RBP2, which is related to RIM-binding protein 1 (RBP1)/PRAX-1 (Galiegue et al., 1999; Wang et al., 2000). We therefore designated the novel protein encoded by the cloned cDNAs as cRBP2, for chicken RBP2.
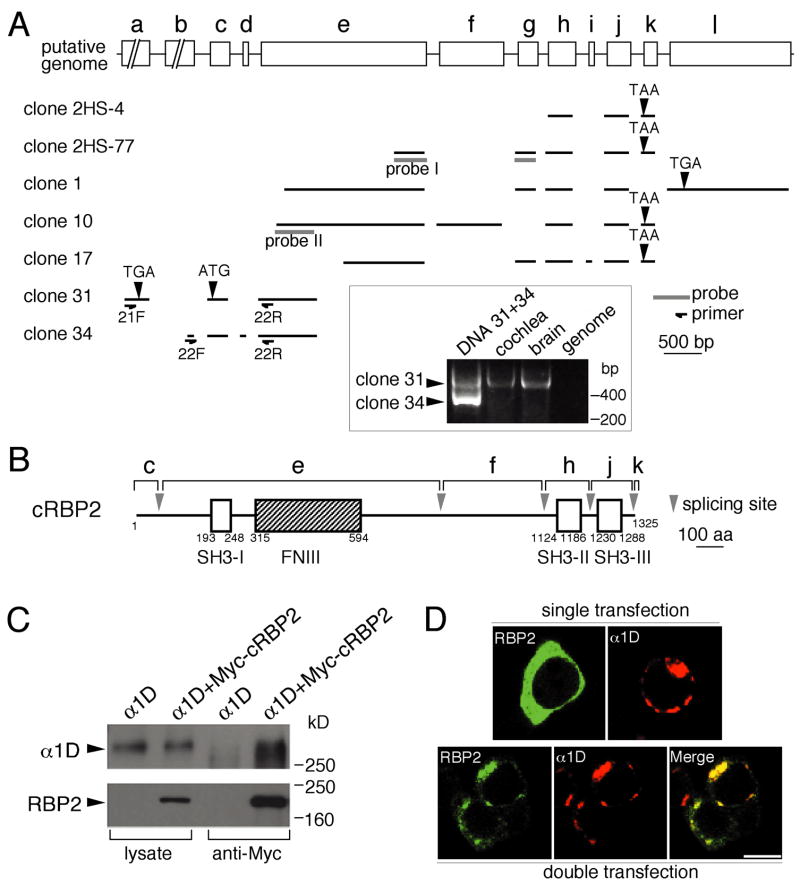
Figure 1. Interaction of RBP2 with the Ca2+ channel α1D subunit.
(A) Cloning of RBP2 cDNAs and deduced gene organization. The cDNA clones 2HS-4 and 2HS-77 encode polypeptides that interact with the carboxyl terminus of the α1D channel subunit in a yeast two-hybrid screen. A 5′ probe (probe I) from 2HS-77 was used to isolate clones 1, 10, and 17 from a cochlear cDNA library. An additional round of screening using a probe corresponding to the 5′ part of clone 10 (probe II) led to the cloning of cDNAs 31 and 34 from a brain library. Sequencing of these clones revealed the occurrence of alternative splicing in the RBP2 gene. The minimal number of exons and their relative positions are indicated (exons a to l). Inset: PCR analysis indicates that the mRNA corresponding to clone 31, but not that associated with clone 34, is abundant in brain and cochlea.
(B) RBP2 protein structure. A full-length cDNA was obtained by ligating clones 10 and 31. The positions of Src homology domain 3 motifs and repeat of three contiguous fibronectin type III domains in the deduced RBP2 protein are indicated as respectively SH3 and FNIII.
(C) Coimmunoprecipitation of RBP2 and α1D from transfected tsA201 cells. First and second lanes: In Western blots of the lysates from transfected cells expressing α1D alone or α1D plus a myc-tagged form of RBP2, α1D was detected by use of specific affinity-purified antibodies and myc-tagged RBP2 was identified by anti-myc antibodies. Third and fourth lanes: α1D was precipitated by anti-myc antibodies when it was coexpressed with myc-RBP2 but not when it was expressed alone.
(D) Subcellular colocalization of myc-tagged RBP2 and α1D in transfected tsA201 cells. Expressed alone, RBP2 (fluorescein, green) has a diffuse cytoplamic distribution (upper left panel) and α1D (Texas red, red) is clumped (upper right panel). When RBP2 is coexpressed with α1D, its distribution changes and largely overlaps that of α1D, yielding yellow in the merge panel. Scale bar: 10 μm.
To obtain a full-length cDNA encoding RBP2, we sequentially screened chicken cochlear and brain cDNA libraries (Fig. 1A). Two clones (clones 31 and 34) corresponding to divergent 5′ ends of RBP2 cDNA were isolated from the brain library. RT-PCR analysis of cDNA from the chicken’s cochlea and brain indicated that the sequence corresponding to clone 31, which contained an ATG codon downstream of an in-frame stop codon, is more strongly expressed in both tissues than that corresponding to clone 34 (Fig. 1A, inset). We therefore used clone 31 to assemble a full-length cDNA for cRBP2 (GenBank accession number AY072908).
The deduced structure of cRBP2 (Fig. 1B) includes an initial Src homology 3 (SH3) domain (SH3-I), three contiguous fibronectin type III domains (FNIII), and two additional SH3 domains (SH3-II and SH3-III). This overall organization is identical to that of rat RBP1/PRAX-1 (Galiegue et al., 1999; Wang et al., 2000), to which chicken RBP2 is 38% identical. Chicken RBP2 is 84% identical to the partial rat RBP2 sequence (Wang et al., 2000) over the range of overlap.
By analyzing the sequences of all cDNAs isolated, we deduced that the RBP2 gene contains at least twelve exons (Fig. 1A, top panel) and that alternative splicing occurs from this gene. The three SH3 domains and three fibronectin motifs seem to be conserved in the splice variants that are produced by this gene.
We next verified that RBP2 forms a protein complex with the α1D subunit of Ca2+ channels upon expression in mammalian cells (Fig. 1C). tsA201 cells were transiently transfected with the α1D subunit in the presence or absence of a myc-tagged form of RBP2 and the cell lysates were used for immunoprecipitation. The anti-myc tag antibodies precipitated α1D when myc-RBP2 was coexpressed (Fig. 1C). No precipitation of the α1D subunit by anti-myc antibodies was observed from a lysate of cells expressing only α1D. Moreover, the subcellular distribution of RBP2 was strikingly modified upon coexpression with α1D (Fig. 1D). When expressed alone, RBP2 displayed a diffuse cytoplasmic distribution, whereas α1D was clustered. When coexpressed, the two proteins showed an overlapping, clustered distribution. These results indicate that RBP2 and α1D interact when they are coexpressed.
Colocalization of RBPs and α1D
We next determined the cellular and subcellular localization of RBP2 in the brain, cochlea, and retina. When used in Western blot analysis, affinity-purified antibodies directed against the carboxyl-terminal region of RBP2 detected four proteins in the brain (Fig. 2A). The full-length RBP2 expressed in tsA201 cells is approximately equal in size to the largest of these. Because antibodies against RBP2 also recognize the carboxyl-terminal domain of RBP1 (not shown), some of the proteins detected in the brain may have represented RBP1 and its splice variants. The three smaller proteins may have corresponded to the products of alternatively spliced mRNAs for both genes lacking some of the exons delineated in Fig. 1A. We therefore refer to the proteins recognized by anti-RBP2 antibodies as RBPs.
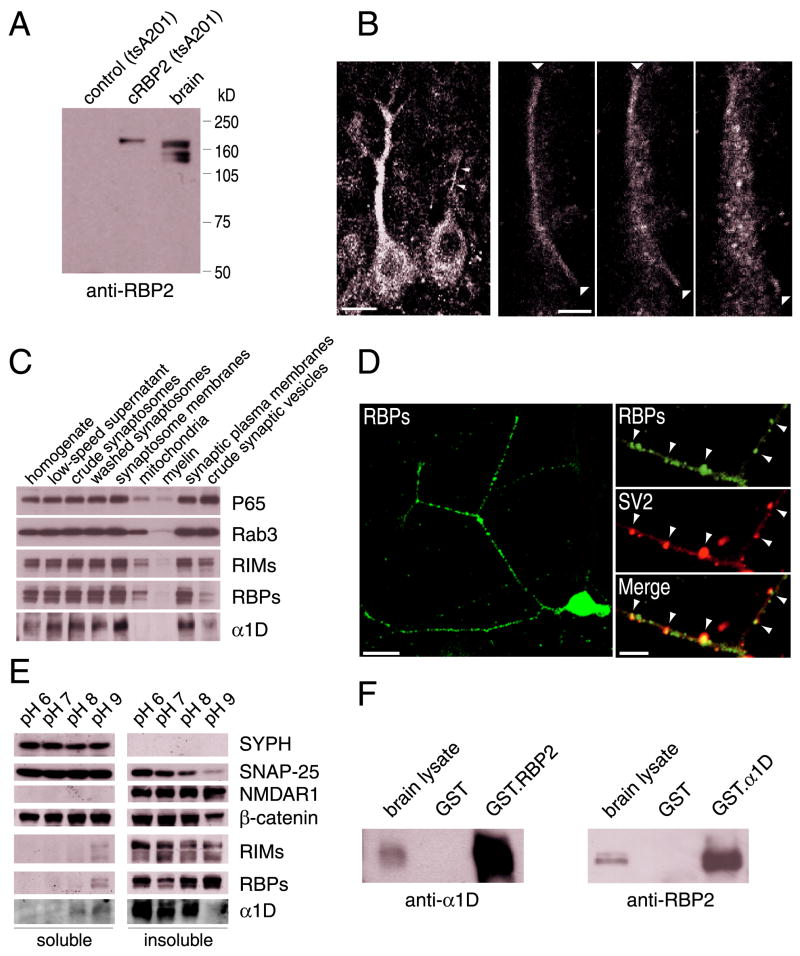
Figure 2. Distribution of RBPs in chicken brain and pulldown of native RBPs and Ca2+ channel α1D subunit.
(A) Identification of RBPs in adult brain by Western blot analysis with affinity-purified antibodies raised against RBP2. Proteins from mock- and RBP2-transfected tsA201 cells were analyzed as negative (first lane) and positive (second lane) controls.
(B) Immunohistochemical localization of RBPs in chicken Purkinje cells. Scale bar: 10 μm. Right panels: serial confocal images showing the RBP2 immunoreactivity of a presynaptic climbing fiber (arrowheads) as well as that of a postsynaptic Purkinje cell dendrite. Scale bar: 3 μm.
(C) Subcellular distribution of α1D and RBPs. Subcellular fractions were prepared from adult chicken brains as described in Experimental Procedures.
(D) RBP2 immunoreactivity in cultured dorsal-root-ganglion neurons from the chicken. Scale bar: 15 μm. Right panels: double labeling with antibodies against RBP2 and the vesicle protein SV2. Scale bar: 3 μm.
(E) Solubility analysis of synaptic proteins. After chicken-brain synaptosomes had been extracted with 1% Triton X-100 at the indicated pHs, equal volumes of soluble and insoluble material were analysed by Western blotting.
(F) Pulldown of native RBPs and α1D. The carboxyl-terminal domains of RBP2 and α1D were expressed as GST fusion proteins, immobilized on gluthatione-Sepharose beads, and used for affinity chromatography. Left panel: Pulldown of α1D by GST·RBP2 fusion protein. Right panel: Pulldown of RBPs by GST·α1D fusion protein. Solubilized synaptic proteins (brain lysate lanes) were analyzed as positive controls; GST alone (GST lanes) provided a negative control.
When used for immunohistochemistry, anti-RBP2 antibodies labeled the chicken’s brain extensively and almost uniformly, indicating a wide distribution of the RBPs (not shown). The labeling was principally neuronal. In the cerebellum, for example, the cell bodies and dendrites of Purkinje cells and the climbing fibers wrapping the dendrites were highly labeled (Fig. 2B), suggesting both post- and presynaptic localizations of the RBPs.
To further characterize the localization of RBPs, we prepared subcellular fractions of brain proteins and probed them with antibodies directed against synaptotagmin/P65, Rab3, RIM1, RBP2, and α1D (Fig. 2C). As expected, synaptotagmin/P65 was enriched in the synaptic-vesicle fraction (Wang et al., 1997). Rab3 occurred in both the synaptic plasma-membrane and synaptic-vesicle fractions, whereas RIMs were found primarily in the synaptic plasma-membrane fraction (Wang et al., 1997). Immunoreactivities to both RBP2 and α1D occurred in the synaptic plasma-membrane fraction. Synaptic localization of RBPs was then confirmed in vivo by using cultured neurons isolated from chicken dorsal root ganglia (Fig. 2D). In these neurons, RBP2 immunoreactivity partially overlaps the SV2 immunoreactivity that labels the synaptic areas (Boudin et al., 2000). To separate presynaptic particles and postsynaptic densities by centrifugation (Phillips et al., 2001), we next extracted brain synaptosomes with 1% Triton X-100 at different pH values. SNAP-25 was present in the pellet at the lowest pH values but was extracted at pH values over 7 as expected for a protein present in the presynaptic specialization (Fig. 2E). Consistent with its linkage to postsynaptic densities, NMDA receptor type 1 (NMDAR1) was found in the insoluble fraction at both low and high pH. As expected, β-catenin, which is present on both pre- and postsynaptic membranes, was found in soluble and insoluble fractions; the vesicle protein synaptophysin was found exclusively in the soluble fraction.
As previously observed (Phillips et al., 2001), the presynaptic proteins RIMs (Fig. 2E) and bassoon (not shown) were insoluble at all pH values tested, indicating that these proteins remain associated with postsynaptic densities under conditions in which the presynaptic particles are extracted. In these experimental conditions, we found that the RBPs distribution was identical to that of RIMs and bassoon (Fig. 2E). The fact that RIM2 and RBP2 expressed in tsA201 cells are completely soluble at all pH values over 6 (not shown) rules out insolubility as an intrinsic property of these proteins, and strongly suggests that RBPs and RIMs belong to a same insoluble presynaptic complex in the brain. Finally, we found that α1D was completely extracted at a pH over 8 (Fig. 2E), a result compatible with the partially presynaptic localization of this channel subunit in the brain.
Because RBPs and α1D occur together in the synaptic plasma-membrane fraction, we solubilized this fraction and used it for GST-pulldown assays of native proteins. A GST fusion protein containing the last two SH3 domains of RBP2 quantitatively precipitated the α1D subunit expressed in the brain (Fig. 2F). Conversely, a GST fusion protein including the cytoplasmic carboxyl terminus of α1D precipitated native RBPs.
In the retina, L-type Ca2+ channels containing α1D and α1F subunits mediate neurotransmission at the ribbon synapses of photoreceptors (Morgans, 1999; Strom et al., 1998). We therefore examined the distribution of RBP2 in the chicken’s retina and compared it to that of α1D. Whereas α1D immunoreactivity was specifically detected in the outer plexiform layer, the distribution of RBPs was far broader (Fig. 3A). RBPs were expressed in the outer nuclear layer, outer plexiform layer (arrowheads), and ganglion cell layer. To examine whether α1D and RBPs are colocalized in the outer plexiform layer, we performed double-immunolabeling using an antibody against SV2, a presynaptic protein specifically expressed at the active zones of ribbon synapses (Yang et al., 2002). The immunoreactivities of α1D and RBPs perfectly overlapped that of SV2 (Fig. 3B).
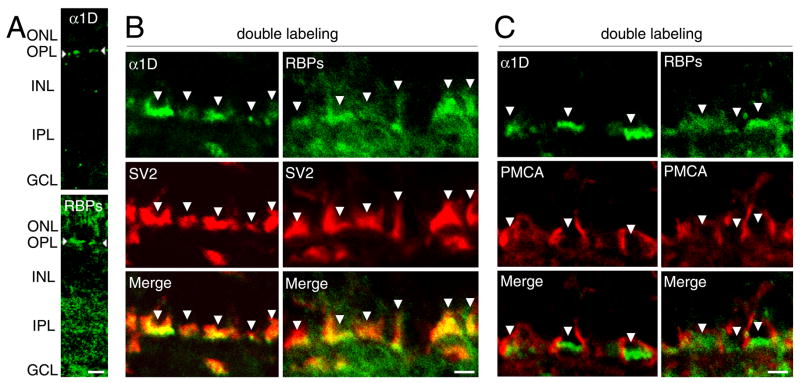
Figure 3. Distribution of RBPs and Ca2+ channel α1D subunit in retinal neurons.
(A) The expression of α1D (upper panel) and RBPs (lower panel) in the chicken’s retina. ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer. Scale bar: 10 μm.
(B) Colocalization of α1D subunit and RBPs in ribbon synapses of photoreceptors. The chicken’s neural retina was immunolabeled with anti-α1D or anti-RBP2 (fluorescein, green) and with anti-SV2 (Texas Red, red). Scale bar: 2 μm.
(C) Distinct distributions of α1D and RBPs from that of plasma-membrane Ca2+-ATPase (PMCA) in photoreceptors. The retina was immunolabeled with anti-α1D or anti-RBP2 (fluorescein, green) and with anti-PMCA (Texas Red, red). Scale bar: 2 μm.
To further confirm the presynaptic distribution of α1D and RBPs, we conducted double-immunolabeling using an antibody against Ca2+-ATPase (PMCA), a protein expressed along the lateral membranes of photoreceptor terminals but not at active zones (Morgans et al., 1998). There was no overlap between the distribution of PMCA and that of α1D or RBPs (Fig. 3C). Therefore, α1D is colocalized with RBPs at the presynaptic active zones of ribbon synapses in photoreceptors. The RBP immunoreactivity observed beneath photoreceptors likely belongs to the postsynaptic processes of bipolar or horizontal cells. Immunohistochemistry also suggested that α1D is colocalized with RBPs at the presynaptic active zones of hair cells (not shown).
Characterization of the α1D-RBP2 interaction
To identify the molecular domains through which RBP2 and α1D interact, we conducted yeast two-hybrid and GST-pulldown assays with deletion and point mutants (Fig. 4). For the pulldown assays, glutathione-Sepharose beads charged with GST fusion proteins were incubated either with lysates of tsA201 cells expressing the full-length α1D subunit or with solubilized synaptic-membrane proteins. Bound proteins were analyzed by Western blotting with anti-α1D or anti-RBP2 antibodies.
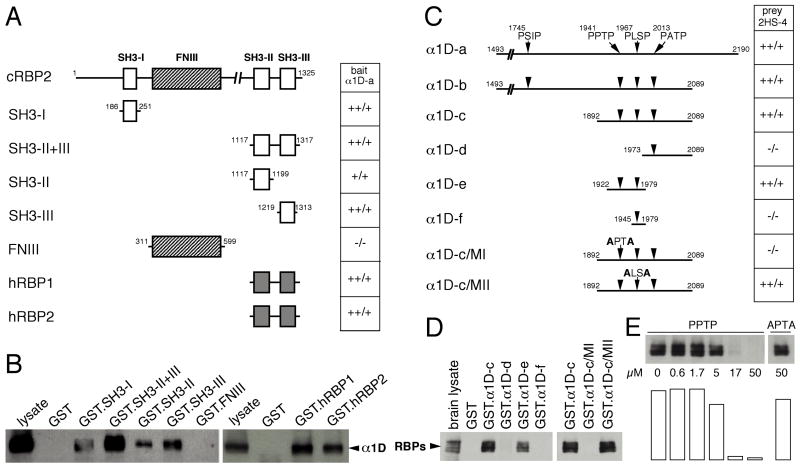
Figure 4. Characterization of the interaction sites of RBPs and α1D by two-hybrid and GST-pulldown experiments.
(A) In a two-hybrid assay with the carboxyl terminus of α1D as a bait, five chicken RBP2 fragments and the carboxyl-terminal portions of human RBP1 and RBP2 were tested. The interactions were scored as the ratio of β-galactosidase activity to His prototrophy. SH3: Src homology 3 domain, FNIII: fibronectin type III repeat.
(B) For GST-pulldown assays, the RBP2 fragments used in A were transferred to a GST plasmid to produce the corresponding GST fusion proteins in bacteria. These proteins were immobilized on gluthatione-Sepharose beads, then incubated with a cell lysate prepared from α1D-transfected tsA201 cells. Bound proteins were analyzed by Western blotting with anti-α1D antibodies.
(C) In a two-hybrid assay with the carboxyl-terminal region of RBP2 as a bait, five α1D fragments and two additional fragments containing point mutations were tested. In the α1D-c/MI fragment, the PPTP motif (residues 1941-1944) had been altered to APTA; in α1D-c/MII, the PLSP motif (residues 1967-1970) had been replaced by ALSA. Interactions were scored as in A.
(D) In GST-pulldown assays, the α1D fragments employed in C were transferred to a GST plasmid to produce the corresponding GST fusion proteins in bacteria. These proteins were immobilized on gluthatione-Sepharose beads, then incubated with solubilized synaptic-membrane proteins from adult chicken brain. Bound proteins were analyzed by Western blotting with anti-RBP2 antibodies.
(E) In a competitive pulldown assay, the GST·α1D-c fusion protein was immobilized on glutathione-Sepharose beads, then incubated with solubilized synaptic-membrane protein from adult chicken brain in the presence of increasing concentrations of peptide. The PPTP peptide was an 18-mer corresponding to residues 1933-1950 of α1D. The APTA peptide was identical to PPTP except that the prolines 1941 and 1944 had been replaced by alanines. Signals were quantified with NIH Image software and are represented in arbitrary units.
In the two-hybrid system, the carboxyl-terminal portion of α1D (α1D-a) interacted with each of the three SH3 domains of RBP2, but not with the fibronectin type III repeat (Fig. 4A). In addition, fragments of human RBP1 and RBP2 corresponding to the last two SH3 domains interacted with α1D. GST-pulldown assays using the full-length α1D subunit gave exactly the same results (Fig. 4B), indicating that the SH3 domains of RBP family members are necessary for interaction with α1D.
We next identified the RBP-interaction site of α1D. This channel subunit contains at its carboxyl terminus the sequence ITSL, a potential site of interaction for PDZ domains. Because a deletion mutant (α1D-b) lacking this site nevertheless bound RBP2, this motif is not essential for interaction.
SH3 domains usually interact with proline-rich domains (reviewed in Mayer, 2001), four of which occur near the carboxyl terminus of α1D. The α1D-c construct lacking the first PXXP motif was able to bind to RBP2. Further deletions of the second and third PXXP motifs abolished this interaction (α1D-d, Fig. 4C and 4D). The construct α1D-e, which contained only the second and third PXXP motifs, clearly bound to RBP2. By contrast, α1D-f, a construct that retained only the third PXXP motif, no longer associated with RBP2. To confirm that the second PXXP motif is responsible for the interaction, we mutated the second (α1D-c/MI) or the third (α1D-c/MII) PXXP motif by replacing the two proline residues with alanines (Fig. 4C). RBP2 bound to α1D-c/MII, but not to the α1D-c/MI mutant. The second motif, PPTP, is therefore the site at which α1D interacts with the SH3 domains of RBPs.
An 18-meric peptide corresponding to the PPTP motif and its flanking sequences was then used to inhibit the binding between GST·α1D-c and native RBPs. The inhibition was concentration-dependent, with 50% inhibition observed at a peptide concentration of around 10 μM (Fig. 4E). Because the interactions between GST·α1D-c and the different RBP proteins found in the brain were similarly inhibited, the different RBPs probably display a comparable affinity for α1D. As expected, a control peptide with a mutated PXXP motif, APTA, had no inhibitory effect on the interaction.
In addition to the four PXXP motifs near the carboxyl terminus, the α1D subunit contains two additional PXXP motifs in intracellular loop II/III and one in intracellular loop III/IV. In two-hybrid experiments, however, neither loop bound to the SH3 domains of RBP2 (not shown). α1D can therefore interact with any of the three SH3 domains of RBP2, and probably those of RBP1, through a unique PXXP sequence near its carboxyl terminus. Two major classes of ligands for SH3 domains have been identified (Mayer, 2001). Class I ligands have the general consensus +XΦPXΦP whereas class II ligands display the consensus sequence ΦPXΦPX+, in which + is a basic residue, usually arginine, X is any amino acid, and Φ is a hydrophobic residue. The α1D motif, RLLPPTP, fits the consensus sequence for class I ligands, differing only by a non-hydrophobic threonine residue in the sixth position.
In the yeast two-hybrid system, there was no apparent interaction between the SH3 domains of RBPs and the RRQPPTP motif of the type 1 GABAB receptor. In addition, α1D was not pulled down by GST fusion proteins containing SH3 domains from proteins as different as the p47phox subunit of NADPH oxidase, p85 subunit of phosphatidyl-inositol 3-kinase, adapter molecule Crk, amphiphysin I, and p21ras GTPase-activating protein (not shown). These observations provide strong support for a specific interaction between the SH3 domains of RBPs and the motif that we have identified in α1D.
RBPs potentially interact with other Ca2+-channel α1 isoforms
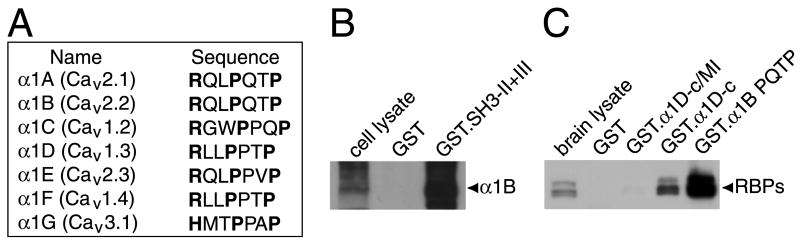
The distribution of RBPs in the brain is far broader than that of α1D (Hell et al., 1993). Because N- and P/Q-type Ca2+ channels are abundant and more widely distributed in the brain than L-type channels (Westenbroek et al., 1992; Westenbroek et al., 1995), α1 subunits other than α1D may interact with RBPs. To test this hypothesis, we first searched protein databases for other α1 subunits that contain a motif similar to the SH3 domain-binding sequence of α1D. We determined that this motif—including the unusual threonine found in α1D—is conserved among α1A, α1B, and α1F subunits (Fig. 5A). α1E lacks the threonine residue in the sixth position but otherwise accords with the α1D sequence. We therefore examined the interaction between RBPs and the α1B subunit of the N-type Ca2+ channel (Fig. 5B and 5C).
Figure 5. Interaction of RBPs with various Ca2+-channel α1 subunit isoforms in the brain.
(A) Voltage-gated Ca2+ channel α1 subunits contain sequences similar to the RBP2-binding site of α1D. Accession numbers: α1A, NP_075461; α1B, NP_000709; α1C, CAA84353; α1D, AF027602; α1E, XP_001815; α1F, NP_005174; α1G, O43497.
(B) The α1B subunit was expressed in tsA201 cells, then precipitated by a GST fusion protein (GST·SH3-II+III) containing the last two SH3 domains of RBP2. α1B was detected with specific antibodies.
(C) Native brain RBPs were precipitated by a GST fusion protein (GST·α1BPQTP) containing the RQLPQTP motif of rat α1B and its flanking sequences (residues 2158-2221). RBPs were detected with anti-RBP2 antibodies.
A GST fusion protein containing the last two SH3 domains of RBP2 quantitatively precipitated a full-length α1B subunit expressed in tsA201 cells (Fig. 5B). Furthermore, native brain RBPs were precipitated by a GST fusion protein containing the RQLPQTP motif of α1B and its flanking sequences (Fig. 5C). Taken together, these results suggest that RBPs can interact with α1B and perhaps with other α1 isoforms in the brain.
SH3 domains of RBPs interact with RIMs
RBP1 and a fragment of RBP2 were first identified by their capacity to interact with RIM1, a synaptic protein involved in neurotransmitter release (Wang et al., 2000). The authors of the original study suggested that the interaction between the RBPs and RIM1 resulted from binding of a SH3 domain of RBPs to a PXXP motif located between the two C2 domains of RIMs (Fig. 6A). Consistent with this suggestion, we found that a GST fusion protein containing the last two SH3 domains of RBP2 specifically precipitated native RIMs from a brain lysate (Fig. 6B). Although the antibodies used for this Western blot analysis were directed against RIM1, they were not specific but cross-reacted with the closely related protein RIM2 (not shown). The multiple proteins detected by these antibodies may therefore have corresponded to RIM1, RIM2, and splice variants of either or both proteins (Ozaki et al., 2000; Wang et al., 2000).
Figure 6. Characterization of the RBP-RIM interaction.
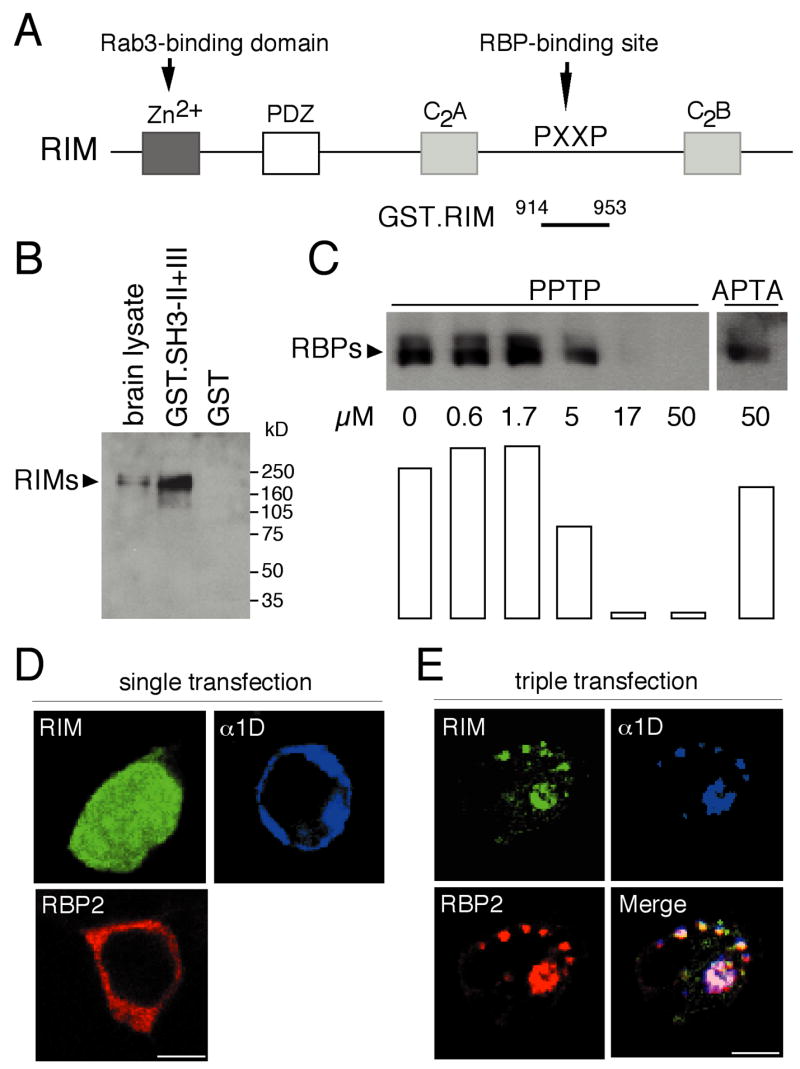
(A) Structure of Rab3-interacting molecules (RIMs).
(B) In a GST-pulldown assay, a GST fusion protein containing the carboxyl terminus of RBP2 was immobilized on glutathione-Sepharose beads, then incubated with solubilized brain proteins. Bound proteins were analyzed by Western blotting with anti-RIM1 antibodies.
(C) A GST fusion protein containing residues 914-953 of RIM2 was immobilized on glutathione-Sepharose beads and incubated with solubilized brain proteins in the presence of various concentrations of PPTP peptide or with 50 μM of APTA peptide. Bound proteins were analyzed by Western blotting with anti-RBP2 antibodies. Signals were quantified by using NIH Image software and are represented in arbitrary units.
(D) Single transfection in tsA201 cells of RIM-GFP (GFP, green), α1D (Cy-5, blue), or myc-tagged RBP2 (Texas Red, red) results in differing distributions.
(E) After cotransfection of the three clones, RIM, α1D, and RBP immunoreactivities are colocalized. Scale bar: 3 μm.
We next produced a GST fusion protein with a fragment of RIM2 containing a specific PXXP motif. This fusion protein precipitated RBPs from a brain lysate (Fig. 6C). The PXXP motif of RIM1, RQLPQVP, closely resembles the RQLPQLP motif of RIM2. Because these sequences are found at equivalent positions in the two proteins, the previously reported interaction between RIM1 and RBP2 is also likely to be mediated by this PXXP site. The interaction of brain RBPs with RIM2 was inhibited by a peptide corresponding to the RBP2-interacting site of α1D (Fig. 6C). This result confirms that the SH3 domains of RBPs mediate binding to the RIMs. Moreover, the SH3 domains of RBP1 and RBP2 that bind to RIM1 and RIM2 can also interact with α1D. That each RBP possesses three SH3 domains raises the possibility that RBPs may interact simultaneously with RIM and α1. In the absence of RBP2, α1D and a RIM2-GFP fusion protein did not occur together upon heterologous expression in cotransfected cells (not shown). When the full-length RBP2 and α1 proteins were coexpressed with RIM2-GFP, however, the three were clearly colocalized (Fig. 6D and 6E). RBP2 is therefore able to bind RIM and α1D simultaneously.
Inhibition of RBP interactions increases hormone secretion in PC12 cells
To inquire whether RBPs are involved in Ca2+-dependent exocytosis, we used a human growth hormone (GH) coexpression assay with PC12 cells. After GH and a polypeptide to be tested had been cotransfected, the cells were stimulated with a high K+ concentation in the presence of Ca2+. The elevated K+ concentration caused membrane depolarization, opening of voltage-gated Ca2+ channels, entry of Ca2+, and finally vesicle fusion and GH release. PC12 cells bear N-type Ca2+ currents mediated by α1B channel subunits (Liu et al., 1996) and express both RBPs (not shown) and RIM2 (Ozaki et al., 2000). These cells therefore constitute a useful model system in which to test the physiological function of interactions between RBPs and both α1D and RIMs.
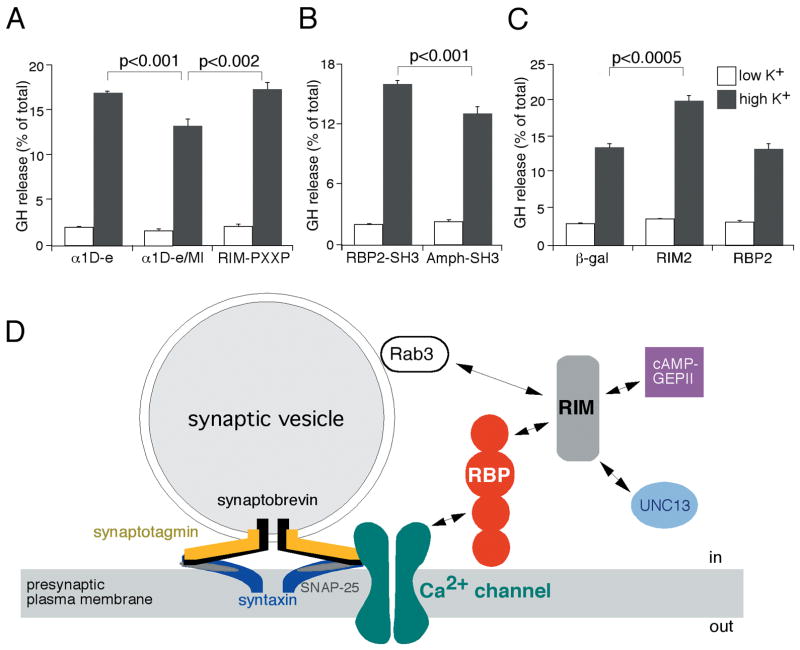
Overexpression of the α1D-e fragment of α1D that associates with RBPs would be expected to inhibit the interaction of native RBPs with Ca2+-channel α1 subunits as well as with RIMs. An α1D-e/MI mutant of α1D-e that cannot interact with RBP2 was chosen as a negative control. cDNAs encoding these fragments were fused to the cDNA encoding DsRed fluorescent protein and transfected into PC12 cells expressing GH. By monitoring the fluorescence produced by the DsRed fusion proteins, we verified that the transfection efficiencies and expression levels were identical for the two proteins (not shown). We then measured the secretion of GH induced by high-K+ treatment as well as the basal GH secretion in a control solution (Fig. 7A).
Figure 7. Enhanced secretion in PC12 cells owing to overexpression of fusion proteins that inhibit RBP interactions.
(A) Expression of α1D-e increases the depolarization-activated GH secretion in PC12 cells by comparison with α1D-e/MI, which differs only by two residues in the RBP2-binding site. A similar effect was observed by expressing the RBP-binding site of RIM2 (RIM-PXXP, residues 914-953).
(B) Expression of the third SH3 domain of RBP2, which is a RIM- and α1D-binding site (RBP2-SH3, residues 1210-1317), specifically enhances stimulated GH secretion when compared to the expression of the SH3 domain of amphiphysin I (Amph-SH3, residues 588-695), which is not able to interact with RBP and α1 subunits.
(C) Expression of full-length RIM2, but neither full-length RBP2 nor the negative control β-galactosidase, enhances stimulated GH secretion.
(D) By virtue of their multiple SH3 domains, RBPs may act as bifunctional linkers between RIMs and voltage-gated Ca2+ channels, thus forming a physical connection between the priming and fusion apparatus constituted by the SNARE complex and the vesicles tethered by RIM and Rab3 at the presynaptic active zone. The proteins cAMP-GEPII and UNC-13 also interact with RIM.
The control construct α1D-e/MI altered neither the basal rate of GH secretion nor that induced by depolarization in comparison with DsRed-mock plasmid (not shown). The construct α1D-e, which does interact with RPBs, also did not affect basal GH secretion. Upon cellular depolarization, however, this construct significantly enhanced GH secretion in comparison to the level achieved with α1D-e/MI (16.7%±0.1% versus 13.1%±0.5%, mean±standard error, n=8 experiments). To confirm this result, we performed a similar experiment with a DsRed construct (RIM-PXXP) containing the PXXP motif of RIM2, which interacts with the same SH3 domains in RBPs as the PXXP motif found in α1D-e. As expected, this RIM-PXXP construct was able to enhance the stimulated GH secretion (17.1%±0.1%, n=8).
If overexpression of the RBP-binding sites of α1D and RIM2 is able to enhance the stimulated GH exocytosis by inhibiting the interactions of RBPs with α1D and RIMs, overexpression of the α1D- and RIM-interacting sites of RBP2 is expected to produce the same effect. To test this hypothesis, we overexpressed the third SH3 domain of RBP2 (RBP2-SH3) in PC12 cells (Fig. 7B). The SH3 domain of amphiphysin I (Amph-SH3) was chosen as a negative control, because its GST fusion protein does not interact with α1D and the neurons of amphiphysin I knock-out mice release glutamate normally (Di Paolo et al., 2002). As anticipated, the expression of RBP2-SH3 was associated with a significant increase of stimulated GH secretion when compared to the expression of Amph-SH3 (Fig. 7B; 15.9%±0.3% versus 13.0%±0.5%, n=8).
Finally, we examined the effects of expressing full-length RIM2 and RBP2 (Fig. 7C). RIM2 significantly enhanced stimulated GH secretion (19.8%±0.9% versus 13.4%±0.6% for the negative control β-galactosidase, n=8), whereas RBP2 had no effect (13.1%±0.6%).
Discussion
Neurotransmitter release is marked by its spatial restriction to synaptic active zones and its efficiency even at high action-potential frequencies. To ensure the coordination of synaptic-vesicle trafficking and exocytosis, the processes of vesicle docking, priming, and fusion are likely to be coupled at the molecular level. The many proteins known to be involved in neurotransmitter secretion have indeed been found to engage in a complex network of interactions.
We have shown that RBPs interact with synaptic RIMs and voltage-gated Ca2+ channels. RIM1 and RIM2 are active zone-specific proteins, each of which contains a PDZ domain, two C2 domains near the carboxyl terminus, and a pair of Cys4 zinc fingers near the amino terminus (Wang et al., 1997). A RIM is associated with the plasma membrane, presumably through its PDZ or C2 domain, whereas its amino-terminal regions interacts with Rab3 in the GTP-bound form (Wang et al., 1997). Rab3 is involved in synaptic-vesicle trafficking and interacts with the vesicle membrane when bound to GTP (Geppert and Südhof, 1998). RIMs may therefore recruit vesicles to the active zone in a tethering reaction. Overexpression of the amino-terminal domain of RIM1, which is expected to suppress the interaction of RIMs with Rab3, increases stimulated secretion in PC12 cells (Wang et al., 1997). This result suggests that the interaction of Rab3 with RIMs limits the number of vesicles that are released during an action potential. In addition, RIMs interact in secretory cells with cAMP-GEFII, a cAMP sensor (Ozaki et al., 2000). This interaction mediates cAMP-induced, PKA-independant Ca2+-dependent secretion.
Voltage-gated Ca2+ channels interact with the SNARE complex that constitutes the fusion machinery (Atlas, 2001; Catterall, 1999). In neurons, α1 subunits, more specifically α1B for N-type channels and α1A for P/Q-type channels, interact with syntaxin, SNAP-25, and synaptotagmin. These interactions provide an effective association between Ca2+-entry and vesicle-release sites that ensures the rapid triggering of neurotransmitter release when an action potential invades the nerve terminal. Similar interactions have been reported to occur in endocrine cells with the α1C and α1D subunits of L-type channels, which are important for the fast stimulated secretion of hormones.
RBP1 and RBP2 form a novel class of proteins displaying three SH3 domains and three contiguous fibronectin type III domains. Because the SH3 domains are more closely related to one another than to any other SH3 sequences deposited in protein databases, these domains probably recognize specific PXXP motifs. We have established that these SH3 domains can bind to RQLPQL/VP, a motif found in RIM1 and RIM2, to RLLPPTP, a motif found in α1D, and to RQLPQTP, a motif found in α1B and α1A. The binding of RBPs to α1D or to RIMs can be inhibited by a synthetic peptide corresponding to the RBP-interacting site of α1D. The half-maximal concentration for this inhibition is near 10 μM, a value that accords with the affinities of specific ligands for other SH3 domains (Mayer, 2001).
In the brain, RBPs have a very wide distribution and are not specifically localized to presynaptic active zones. Like SNAP-25 and syntaxin (Garcia et al., 1995), RBPs also occur postsynaptically in dendrites and cell bodies. This distribution argues against an exclusive role of the RBPs in the targeting of RIMs or Ca2+ channels to the presynaptic membrane. A more plausible function for RBPs in the presynaptic area is as a scaffold for the association between RIMs and voltage-gated Ca2+ channels. According to this model, RBPs act as bifunctional linkers that interact simultaneously with RIMs, which bind Rab3-GTP present on vesicles, and with Ca2+ channels, which are associated with the SNARE complex that constitutes the vesicle-fusion machinery. RBPs could therefore provide a molecular coupling between the vesicle-tethering and the priming-fusion apparatus (Fig. 7D). The domain structure of RBPs—three SH3 domains and three fibronectin III motifs—further suggests that RBPs provide the exocytotic machinery with other regulatory proteins through their free domains. A similar role as a link between the vesicle-tethering and -release apparatus has been proposed recently for UNC-13, which interacts with both RIM1 and syntaxin (Betz et al., 2001). It has also been suggested that RIM1 is important for vesicle priming (Koushika et al., 2001; Lloyd and Bellen, 2001). The binding of active Rab3-GTP to the synaptic vesicle may activate RIMs. Once released from Rab3 after GTP hydrolysis, the activated RIMs may bind and stimulate UNC-13, which in turn dissociates the complex of UNC-18 and syntaxin by promoting a conformational change of the syntaxin. This “open” syntaxin can form core complex with synaptobrevin and SNAP-25, priming the synaptic vesicle for fusion.
The enhancement of depolarization-induced secretion in PC12 cells by fusion proteins that suppress the associations of RBPs with RIMs and α1 suggests that RBPs may repress RIMs, either directly or through associated proteins. If the interaction of RBPs with RIMs were prevented, more RIMs would be free to activate UNC-13, resulting in an increase of primed vesicles and ultimately in stimulated secretion. Overexpression of RIMs would also be expected to lead to an increase in the number of primed vesicles. The lack of effect of RBP overexpression in PC12 cells may indicate that every native RIM already interacts with an endogenous RBP.
The increase in stimulated secretion could alternatively reflect inhibition of the interaction between RBPs and Ca2+ channels rather than of that between RBPs and RIMs. The expression level and electrophysiological properties of voltage-gated Ca2+ channels are modulated by various auxiliary and accessory subunits (Catterall, 1999). The association of neuronal Ca2+ channels with syntaxin, for example, causes a change in the voltage dependence of channel inactivation (Bezprozvanny et al., 1995). The interaction of RBPs with Ca2+ channels might tonically repress channel activity. Inhibition of this interaction would then result in increased channel activity, more cytoplasmic Ca2+, and ultimately an increase of Ca2+-dependant exocytosis.
RIMs have been suggested to bind directly to the α1B and α1C subunits of voltage-gated Ca2+ channels (Coppola et al., 2001). We failed to observe any interaction between full-length α1B and RIM2 from transfected cells, whereas an association of RBP2 and RIM2 was clear under the same experimental conditions (unpublished results). However, this result does not signify that α1B and RIMs could not interact under other conditions. Such an association would provide additional support for a complex network of interactions between the different components of the exocytotic machinery. Coppola and co-authors reported that this interaction did not occur between RIMs and the α1D subunits that constitute the presynaptic Ca2+ channels in hair cells and photoreceptors. Hair cells lack synaptophysin and synapsin (Safieddine and Wenthold, 1999), which are components of the exocytotic machinery at brain synapses. The unique interaction of RBPs with α1D and RIMs in the hair cell is also a distinctive property of these sensory cells, and may be associated with some of the specific properties of their synaptic transmission.
Experimental Procedures
Yeast two-hybrid assays
Yeast two-hybrid screening was performed using the GAL4 system. DNA encoding a bait consisting of residues 1493-2190 from the cytoplasmic carboxyl terminus of the α1D subunit (Kollmar et al., 1997a, 1997b) in frame with the DNA-binding domain of GAL4 was constructed by PCR and subcloned into the pBDGal4 vector (Stratagene, La Jolla, CA). A sensory epithelial cDNA library was constructed in the pADGal4 prey vector from basilar papillae of late embryonic chickens, then screened with the α1D bait. Positive clones were selected by His prototrophy and assayed for β-galactosidase activity. Doubly positive clones were isolated and characterized by sequencing. From 10 million cDNA clones screened, 20 were recovered and analyzed. Of seven positive cDNAs, two were identical to cDNA 2HS 77-A and five to cDNA 2HS 4-A. These cDNAs encode overlapping regions of RBP2. For the localization of the interacting sites between α1D subunit and RBP2, additional baits of a α1D cytoplamic domain and additional preys of RBP2 were generated by PCR amplification.
Cloning of the full-length RBP2 cDNA and RT-PCR analysis
The complete sequence of RBP2 was determined from partial cDNA clones obtained by screening two cDNA libraries. The 5′ region of clone 2HS 77-A was first amplified by PCR, subcloned into the TA vector (pCR2.1; Invitrogen, Carlsbad, CA), and used as a probe (probe I, Fig. 1A) for screening of a chicken sensory-epithelium library constructed in the HybriZAP vector (Heller et al., 1998). Clones 1, 10, and 17 were isolated. Using a PCR product corresponding to the 5′ terminal region of clone 10 (probe II, Fig. 1A) as a probe, we then screened a chicken brain library (5′-STRETCH, Clontech, Inc., Palo Alto, CA). The positive clones 31 and 34 were sequenced on both strands. To obtain the full-length cDNA for RBP2, we fused clones 10 and 31 and ligated them into the pCMV-myc (Clontech) and pcDNA3.1(+) vectors (Invitrogen).
For RT-PCR experiments, cochleae and brains were dissected from one-week-old chickens of the White Leghorn strain and total RNAs were isolated with Trizol (Gibco BRL, Rockville, MD). cDNAs were synthesized with Superscript II reverse transcriptase (Life Technologies, Grand Island, NY). Oligo-(dT) and random-hexamer primers were independently used to obtain cDNAs, and the two types of cDNA were mixed. The equivalent of 400 ng of total RNA was used for each PCR reaction. The sequences of primers specific to cRBP2 (Fig. 1A) were: 21F, 5′-GTAGACTGCAGAGCTTTCTCG-3′; 22F, 5′-CCTGGAGCCCGTCAGTATC-3′; and 22R, 5′-CATTTTTCCTCTAGCTCTCG-3′.
Antibodies
Polyclonal antibodies were generated against the chicken α1D subunit and RBP2. Rabbit anti-α1D antibodies were raised against a (His)6 fusion protein corresponding to residues 1973-2190 and affinity-purified against an equivalent GST·α1D fusion protein transferred onto nitrocellulose. Rabbit anti-RBP2 antibody was raised against a GST fusion protein corresponding to residues 1100-1325 and affinity-purified using an equivalent (His)6-RBP2 fusion protein. Antisera directed against β-catenin, synaptotagmin/p65, Rab3, and RIM1 were purchased from BD Transduction Laboratories (Lexington, KY). Antisera against α1B, myc, NMDAR1, SNAP-25, synaptophysin, and PMCA were obtained from Affinity Bioreagents (Golden, CO), Alomone Laboratories (Jerusalem, Israel), Clontech, and Santa Cruz Biotechnology (Santa Cruz, CA).
Subcellular fractionation
Subcellular fractions of the chicken brain were prepared essentially as described (Jones and Matus, 1974) and were maintained at −70°C until use. Solubility analysis of synaptic proteins was conducted as described previously (Phillips et al., 2001). Chicken brain synaptosmes were prepared using a one-step preparation method based on the known isopycnic densities of various cellular components, then diluted in ice-cold 0.1 mM CaCl2. An equal volume of solubilization solution (2% Triton X-100, 0.1 mM CaCl2, and 40 mM Tris buffered at various pH values) was added and the samples were mixed and incubated 30 min on ice. The samples were then centrifuged at 10,000 × g. Supernatants were collected and pellets resuspended in an equal volume of 0.1 mM CaCl2. The samples were analyzed immediately after preparation. After subcellular fractions and synaptic proteins had been separated on 4%–20% SDS-polyacrylamide gels, the proteins were transferred to nitrocellulose and the membranes were probed with antibodies.
Production of glutathione-S-transferase (GST) fusion proteins
DNA fragments corresponding to specific regions of chicken α1D subunit and RBP2, human RBP1 and RBP2, rat α1B subunit, and human RIM2 were amplified by PCRs and ligated into the pGEX-4T-1 vector (Amersham Pharmacia Biotech, Piscataway, NJ). Point mutations of α1D-c/MI and α1D-c/MII were created by using the overlap PCR method (Ho et al., 1989). The BL21 strain of Escherichia coli was transformed by the expression vectors and fusion protein expression was induced by the addition of 1 mM IPTG for 3–4 hrs at 37°C. Bacteria were lysed by sonication in PBS containing 1% Triton X-100, 1 mM EDTA, and a cocktail of protease inhibitors (Roche). After centrifugation, lysates were recovered and the fusion proteins were purified on glutathione-Sepharose.
tsA201 cell transfection, coimmunopreciptation, and GST-pulldown assay
Using Lipofectamine (Gibco BRL) according to the manufacturer’s protocol, we transfected tsA201 cells with the full-length α1D subunit inserted in the GW1 vector in the presence or absence of a myc-tagged full-length cRBP2 plasmid. Cells were harvested 48 hr after transfection. After extraction with a lysis solution containing 1% Triton X-100 and 150 mM NaCl in 40 mM Tris-HCl at pH 7.4, proteins were incubated overnight at 4°C with beads conjugated to anti-myc antibodies (Clontech). The beads were washed four times with the same solution. Immunoprecipitated proteins were resolved by SDS-PAGE and probed with anti-α1D antibody.
For pull-down assays, tsA201 cells transfected with full-length α1D subunit, rat α1B subunit, or brain synaptic plasma membranes were solubilized in PBS containing 1% Triton X-100, 2 mM EDTA, and a proteinase inhibitor cocktail (Roche), then centrifuged at 100,000 × g for 15 min at 4°C. One hundred micrograms of solubilized proteins was incubated overnight at 4°C with 30 μl of glutathione-Sepharose beads bound to 5 μg of purified fusion protein. Samples were washed four times at room temperature with 0.05% Triton X-100 in PBS. The materials retained on the beads were eluted with sample-buffer solution and analysed by SDS-PAGE and immunobloting using anti-α1D, anti-α1B, anti-RIM1, or anti-RBP2 antibodies.
Immunohistochemistry
Immunohistochemistry was performed as described previously (Hibino et al., 1997) with two-week-old chickens and adult male C57BL/6 mice. Retinae were isolated after fixation by perfusion with 4% formaldehyde in PBS. Twelve-micrometer-thick cryosections were incubated with antibodies against α1D, RBP2, SV2, or PMCA and treated with fluorescein-conjugated anti-rabbit (α1D and RBP2) or Texas Red-labeled anti-mouse (SV2 and PMCA) antiserum. For immunocytochemistry of tsA201 cells, the transfected cells were incubated with anti-myc and anti-α1D antisera and visualized with respectively fluorescein- and Texas Red- or Cy-5-labeled secondary antibodies. A cDNA encoding residues 914-953 of RIM2 was inserted into the pEGFP-C2 vector (Clontech); after transfection, the product was directly visualized by GFP fluorescence. Images were obtained with a laser-scanning confocal microscope (MRC-1024; Bio-Rad, Hertfordshire, England). The same procedure was applied for immunocytochemistry of cultured neurons. Dorsal root ganglia from embryonic chickens were dissected and neurons were plated and cultured as described (Nishi, 1996). The anti-SV2 antibody was provided by the Developmental Studies Hybridoma Bank (University of Iowa).
Measurements of growth hormone in cultured cells
DNA fragments of α1D-e, α1D-e/MI, RIM2, RBP2, and amphiphysin I were amplified by PCRs and inserted into the pDsRed1-C1 vector (Clontech). PC12 cells were transfected with 0.5 μg of the pXGH5 plasmid (Nichols Institute Diagnostics, San Juan Capistrano, CA) in the presence of 1 μg of DsRed constructs or full-length β-galactosidase, RBP2, and RIM2 expression plasmids. The cells were maintained in low-K+ solution consisting of 145 mM NaCl, 5.6 mM KCl, 2.2 mM CaCl2, 0.5 mM MgCl2, 5.5 mM glucose, 0.5 mM ascorbic acid, and 20 mM HEPES at pH 7.4. The cells were stimulated for 20 min at 37°C by depolarization in a high-K+ solution of identical composition save for the presence of 95 mM NaCl and 56 mM KCl. The concentration of growth hormone was measured with the hGH ELISA kit (Roche).
Acknowledgments
We thank Dr. Terry P. Snutch (University of British Columbia) for the rat α1B cDNA, Dr. Richard Kollmar (Rockefeller University) for the chicken α1D cDNA, Dr. Stefan Heller (Massachusetts Eye and Ear Infirmary) for the basilar-papilla cDNA libraries, Dr. Susumu Seino (Chiba University) for the RIM2 expression plasmid, and Dr. Pietro De Camilli (Yale University) for amphiphysin I cDNA. Dr. Takuya Sasaki (Tokushima University) advised us on subcellular fractionation; the members of our research group offered valuable comments on the manuscript. H. H. was supported by a Human Frontier Science Program grant (LT0318/1999-B) and F. L. by INSERM, NATO, and the Philippe Foundation. This research was supported by grant DC00317 to A. J. H., who is an Investigator of Howard Hughes Medical Institute.
References
- Atlas D. Functional and physical coupling of voltage-sensitive calcium channels with exocytotic proteins: ramifications for the secretion mechanism. J Neurochem. 2001;77:972–85. doi: 10.1046/j.1471-4159.2001.00347.x. [DOI] [PubMed] [Google Scholar]
- Betz A, Thakur P, Junge HJ, Ashery U, Rhee JS, Scheuss V, Rosenmund C, Rettig J, Brose N. Functional interaction of the active zone proteins Munc13-1 and RIM1 in synaptic vesicle priming. Neuron. 2001;30:183–96. doi: 10.1016/s0896-6273(01)00272-0. [DOI] [PubMed] [Google Scholar]
- Bezprozvanny I, Scheller RH, Tsien RW. Functional impact of syntaxin on gating of N-type and Q-type calcium channels. Nature. 1995;378:623–6. doi: 10.1038/378623a0. [DOI] [PubMed] [Google Scholar]
- Boudin H, Doan A, Xia J, Shigemoto R, Huganir RL, Worley P, Craig AM. Presynaptic clustering of mGluR7a requires the PICK1 PDZ domain binding site. Neuron. 2000;28:485–97. doi: 10.1016/s0896-6273(00)00127-6. [DOI] [PubMed] [Google Scholar]
- Catterall WA. Interactions of presynaptic Ca2+ channels and snare proteins in neurotransmitter release. Ann N Y Acad Sci. 1999;868:144–59. doi: 10.1111/j.1749-6632.1999.tb11284.x. [DOI] [PubMed] [Google Scholar]
- Coppola T, Magnin-Luthi S, Perret-Menoud V, Gattesco S, Schiavo G, Regazzi R. Direct interaction of the rab3 effector rim with Ca2+ channels, snap-25, and synaptotagmin. J Biol Chem. 2001;276:32756–62. doi: 10.1074/jbc.M100929200. [DOI] [PubMed] [Google Scholar]
- Corey DP, Dubinsky JM, Schwartz EA. The calcium current in inner segments of rods from the salamander (Ambystoma tigrinum) retina. J Physiol. 1984;354:557–75. doi: 10.1113/jphysiol.1984.sp015393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G, Sankaranarayanan S, Wenk MR, Daniell L, Perucco E, Caldarone BJ, Flavell R, Picciotto MR, Ryan TA, Cremona O, De Camilli P. Decreased synaptic vesicle recycling efficiency and cognitive defects in amphiphysin 1 knockout mice. Neuron. 2002;33:789–804. doi: 10.1016/s0896-6273(02)00601-3. [DOI] [PubMed] [Google Scholar]
- Galiegue S, Jbilo O, Combes T, Bribes E, Carayon P, Le Fur G, Casellas P. Cloning and characterization of PRAX-1. A new protein that specifically interacts with the peripheral benzodiazepine receptor. J Biol Chem. 1999;274:2938–52. doi: 10.1074/jbc.274.5.2938. [DOI] [PubMed] [Google Scholar]
- Garcia EP, McPherson PS, Chilcote TJ, Takei K, De Camilli P. rbSec1A and B colocalize with syntaxin 1 and SNAP-25 throughout the axon, but are not in a stable complex with syntaxin. J Cell Biol. 1995;129:105–20. doi: 10.1083/jcb.129.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geppert M, Südhof TC. RAB3 and synaptotagmin: the yin and yang of synaptic membrane fusion. Annu Rev Neurosci. 1998;21:75–95. doi: 10.1146/annurev.neuro.21.1.75. [DOI] [PubMed] [Google Scholar]
- Hama K. Fine structure of the afferent synapse and gap junctions on the sensory hair cell in the saccular macula of goldfish: a freeze-fracture study. J Neurocytol. 1980;9:845–60. doi: 10.1007/BF01205023. [DOI] [PubMed] [Google Scholar]
- Haynes LP, Evans GJ, Morgan A, Burgoyne RD. A direct inhibitory role for the Rab3-specific effector, Noc2, in Ca2+-regulated exocytosis in neuroendocrine cells. J Biol Chem. 2001;276:9726–32. doi: 10.1074/jbc.M006959200. [DOI] [PubMed] [Google Scholar]
- Hell JW, Westenbroek RE, Warner C, Ahlijanian MK, Prystay W, Gilbert MM, Snutch TP, Catterall WA. Identification and differential subcellular localization of the neuronal class C and class D L-type calcium channel alpha 1 subunits. J Cell Biol. 1993;123:949–62. doi: 10.1083/jcb.123.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller S, Sheane CA, Javed Z, Hudspeth AJ. Molecular markers for cell types of the inner ear and candidate genes for hearing disorders. Proc Natl Acad Sci U S A. 1998;95:11400–5. doi: 10.1073/pnas.95.19.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibino H, Horio Y, Inanobe A, Doi K, Ito M, Yamada M, Gotow T, Uchiyama Y, Kawamura M, Kubo T, Kurachi Y. An ATP-dependent inwardly rectifying potassium channel, KAB-2 (Kir4.1), in cochlear stria vascularis of inner ear: its specific subcellular localization and correlation with the formation of endocochlear potential. J Neurosci. 1997;17:4711–21. doi: 10.1523/JNEUROSCI.17-12-04711.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–9. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Hudspeth AJ, Lewis RS. Kinetic analysis of voltage- and ion-dependent conductances in saccular hair cells of the bull-frog, Rana catesbeiana. J Physiol. 1988;400:237–74. doi: 10.1113/jphysiol.1988.sp017119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DH, Matus AI. Isolation of synaptic plasma membrane from brain by combined flotation-sedimentation density gradient centrifugation. Biochim Biophys Acta. 1974;356:276–87. doi: 10.1016/0005-2736(74)90268-5. [DOI] [PubMed] [Google Scholar]
- Kollmar R, Fak J, Montgomery LG, Hudspeth AJ. Hair cell-specific splicing of mRNA for the alpha1D subunit of voltage-gated Ca2+ channels in the chicken’s cochlea. Proc Natl Acad Sci U S A. 1997a;94:14889–93. doi: 10.1073/pnas.94.26.14889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmar R, Montgomery LG, Fak J, Henry LJ, Hudspeth AJ. Predominance of the alpha1D subunit in L-type voltage-gated Ca2+ channels of hair cells in the chicken’s cochlea. Proc Natl Acad Sci U S A. 1997b;94:14883–8. doi: 10.1073/pnas.94.26.14883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koushika SP, Richmond JE, Hadwiger G, Weimer RM, Jorgensen EM, Nonet ML. A post-docking role for active zone protein Rim. Nat Neurosci. 2001;4:997–1005. doi: 10.1038/nn732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin RC, Scheller RH. Mechanisms of synaptic vesicle exocytosis. Annu Rev Cell Dev Biol. 2000;16:19–49. doi: 10.1146/annurev.cellbio.16.1.19. [DOI] [PubMed] [Google Scholar]
- Liu H, Felix R, Gurnett CA, De Waard M, Witcher DR, Campbell KP. Expression and subunit interaction of voltage-dependent Ca2+ channels in PC12 cells. J Neurosci. 1996;16:7557–65. doi: 10.1523/JNEUROSCI.16-23-07557.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd TE, Bellen HJ. pRIMing synaptic vesicles for fusion. Nat Neurosci. 2001;4:965–66. doi: 10.1038/nn1001-965. [DOI] [PubMed] [Google Scholar]
- Mayer BJ. SH3 domains: complexity in moderation. J Cell Sci. 2001;114:1253–63. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- Mochida S. Protein-protein interactions in neurotransmitter release. Neurosci Res. 2000;36:175–82. doi: 10.1016/s0168-0102(99)00128-5. [DOI] [PubMed] [Google Scholar]
- Morgans CW, Far OE, Berntson A, Wassle H, Taylor WR. Calcium Extrusion from mammalian photoreceptor terminals. J Neurosci. 1998;18:2467–2474. doi: 10.1523/JNEUROSCI.18-07-02467.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgans CW. Calcium channel heterogeneity among cone photoreceptors in the tree shrew retina. Eur J Neurosci. 1999;11:2989–93. doi: 10.1046/j.1460-9568.1999.00719.x. [DOI] [PubMed] [Google Scholar]
- Nishi R. Autonomic and sensory neuron cultures. Methods Cell Biol. 1996;51:249–63. doi: 10.1016/s0091-679x(08)60632-9. [DOI] [PubMed] [Google Scholar]
- Ozaki N, Shibasaki T, Kashima Y, Miki T, Takahashi K, Ueno H, Sunaga Y, Yano H, Matsuura Y, Iwanaga T, et al. cAMP-GEFII is a direct target of cAMP in regulated exocytosis. Nat Cell Biol. 2000;2:805–11. doi: 10.1038/35041046. [DOI] [PubMed] [Google Scholar]
- Phillips GR, Huang JK, Wang Y, Tanaka H, Shapiro L, Zhang W, Shan W, Arndt K, Frank M, Gordon RE, et al. The presynaptic particle web: ultrastructure, composition, dissolution, and reconstitution. Neuron. 2001;32:63–77. doi: 10.1016/s0896-6273(01)00450-0. [DOI] [PubMed] [Google Scholar]
- Platzer J, Engel J, Schrott-Fischer A, Stephan K, Bova S, Chen H, Zheng H, Striessnig J. Congenital deafness and sinoatrial node dysfunction in mice lacking class D L-type Ca2+ channels. Cell. 2000;102:89–97. doi: 10.1016/s0092-8674(00)00013-1. [DOI] [PubMed] [Google Scholar]
- Roberts WM, Jacobs RA, Hudspeth AJ. Colocalization of ion channels involved in frequency selectivity and synaptic transmission at presynaptic active zones of hair cells. J Neurosci. 1990;10:3664–84. doi: 10.1523/JNEUROSCI.10-11-03664.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safieddine S, Wenthold RJ. SNARE complex at the ribbon synapses of cochlear hair cells: analysis of synaptic vesicle- and synaptic membrane-associated proteins. Eur J Neurosci. 1999;11:803–12. doi: 10.1046/j.1460-9568.1999.00487.x. [DOI] [PubMed] [Google Scholar]
- Shirataki H, Kaibuchi K, Sakoda T, Kishida S, Yamaguchi T, Wada K, Miyazaki M, Takai Y. Rabphilin-3A, a putative target protein for smg p25A/rab3A p25 small GTP-binding protein related to synaptotagmin. Mol Cell Biol. 1993;13:2061–8. doi: 10.1128/mcb.13.4.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom TM, Nyakatura G, Apfelstedt-Sylla E, Hellebrand H, Lorenz B, Weber BH, Wutz K, Gutwillinger N, Ruther K, Drescher B, et al. An L-type calcium-channel gene mutated in incomplete X-linked congenital stationary night blindness. Nat Genet. 1998;19:260–3. doi: 10.1038/940. [DOI] [PubMed] [Google Scholar]
- Wang Y, Okamoto M, Schmitz F, Hofmann K, Südhof TC. Rim is a putative Rab3 effector in regulating synaptic-vesicle fusion. Nature. 1997;388:593–8. doi: 10.1038/41580. [DOI] [PubMed] [Google Scholar]
- Wang Y, Sugita S, Südhof TC. The RIM/NIM family of neuronal C2 domain proteins. Interactions with Rab3 and a new class of Src homology 3 domain proteins. J Biol Chem. 2000;275:20033–44. doi: 10.1074/jbc.M909008199. [DOI] [PubMed] [Google Scholar]
- Westenbroek RE, Hell JW, Warner C, Dubel SJ, Snutch TP, Catterall WA. Biochemical properties and subcellular distribution of an N-type calcium channel alpha 1 subunit. Neuron. 1992;9:1099–115. doi: 10.1016/0896-6273(92)90069-p. [DOI] [PubMed] [Google Scholar]
- Westenbroek RE, Sakurai T, Elliott EM, Hell JW, Starr TV, Snutch TP, Catterall WA. Immunochemical identification and subcellular distribution of the alpha 1A subunits of brain calcium channels. J Neurosci. 1995;15:6403–18. doi: 10.1523/JNEUROSCI.15-10-06403.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Standifer KM, Sherry DM. Synaptic protein expression by regenerating adult photoreceptors. J Comp Neurol. 2002;443:275–288. doi: 10.1002/cne.10116. [DOI] [PubMed] [Google Scholar]
- Zenisek D, Steyer JA, Almers W. Transport, capture and exocytosis of single synaptic vesicles at active zones. Nature. 2000;406:849–54. doi: 10.1038/35022500. [DOI] [PubMed] [Google Scholar]
- Zidanic M, Fuchs PA. Kinetic analysis of barium currents in chick cochlear hair cells. Biophys J. 1995;68:1323–36. doi: 10.1016/S0006-3495(95)80305-X. [DOI] [PMC free article] [PubMed] [Google Scholar]