Abstract
The NF-κB/Rel proteins are sequestered in the cytoplasm in association with IκBα. In response to external signals, IκBα is phosphorylated, multi-ubiquitinated, and degraded by proteasomes, thereby releasing NF-κB/Rel proteins to migrate to the nucleus. We have cloned a mouse ubiquitin-conjugating enzyme (mE2), which associates with IκBα. mE2 is homologous to the yeast Ubc9/Hus5 ubiquitin-conjugating enzyme. A transdominant-negative mutant of mE2 had no effect on phosphorylation of IκBα, but delayed its degradation. Correspondingly, tumor necrosis factor-α-inducible NF-κB activity was diminished. We propose that mE2 is directly involved in the ubiquitin conjugation of IκBα, a pivotal step in its degradation pathway.
Keywords: ubiquitination, protein degradation, E2, adenoviral vectors, NF-κB
The transcription factor NF-κB/Rel is a major regulator of the expression of immunomodulatory genes, viral genes, and stress response genes (for review see refs. 1 and 2). The activity of NF-κB/Rel proteins is regulated by their association with a family of inhibitory proteins (IκB family) that bind dimeric NF-κB complexes, mask the nuclear translocation signal, and retain NF-κB in the cytoplasm (1, 3). When cells are stimulated with a variety of inducers such as lipopolysaccharide, tumor necrosis factor (TNF)-α, and interleukin 1, serines-32 and -36 in the N-terminal region of IκBα are rapidly phosphorylated. IκBα is then multi-ubiquitinated at lysines-21 and -22, signaling its degradation through the ubiquitin (Ub)-dependent proteasome pathway (4–12). Degradation of IκBα allows NF-κB to translocate to the nucleus and activate transcription of target genes (1, 13).
The NF-κB signaling pathway requires Ub conjugation for degradation of IκBα. Ub is a highly conserved polypeptide of 76 amino acids. Ub-conjugation is catalyzed in a three-step mechanism (14): (i) the C-terminal glycine of Ub is activated by ATP and Ub-activating enzyme E1; (ii) the Ub-conjugating enzyme (Ubc) E2 and the Ub protein ligase E3 transfer Ub from E1 to the protein targeted for degradation; and (iii) an isopeptide bond is formed between the activated C-terminal glycine of Ub and ɛ-NH2 group of a lysine residue of the target protein. In successive reactions, a multi-Ub chain is synthesized by transfer of Ub to lysine-48 of the previously conjugated Ub molecule (15). The resulting multi-ubiquitinated target protein is recognized as substrate for degradation by 26S proteosome. Generally, E3 proteins provide the specificity for Ub-dependent degradation by recognizing and binding to the target protein. In many cases, however, E2 alone can transfer activated Ub to the target protein. Thus, certain E2 proteins can play an important role in substrate recognition.
We report the identification and molecular cloning of mouse E2 (mE2) cDNA. mE2 is a homologue of the Ubc9, Hus5 Ub-conjugating enzyme family of Saccharomyces cerevisiae and Schizosaccharomyces pombe, respectively. We show that mE2 associates with IκBα in vitro and is directly involved in the signal-induced degradation of IκBα in vivo.
MATERIALS AND METHODS
Cell Culture.
WEHI231, a mouse B cell line was maintained in RPMI medium 1640 supplemented with 10% fetal calf serum, 5 mM glutamine, and 50 μM 2-mercaptoethanol in an atmosphere of 5% CO2 in air humidified at 37°C. 293T, a variant of the human embryonic kidney cell line 293 and HeLa cells, were maintained in DMEM supplemented with 10% fetal calf serum in an atmosphere of 10% CO2, at 37°C.
Yeast Two-Hybrid Screen and cDNA Isolation.
For the yeast two-hybrid screen, the mouse cRel (1–311) was inserted into the pAS2 vector (a gift from S. Elledge, Baylor College of Medicine, Houston, TX; refs. 16–18), which fuses with the GAL4 DNA-binding domain resulting in a plasmid pAS2-cRel-N. WEHI231 mouse B cell cDNA library in pVP16 vector (gift of S. Hollenberg, Fred Hutchinson Cancer Research Center, Seattle, WA) was constructed (19, 20).
For the screen, first, pAS2-cRel-N plasmid was transformed into the Y190 yeast strain (16), and Y190-cRel-N was established. cDNA library plasmids were transformed into Y190-cRel-N, and 1 × 107 transformants were analyzed as described by Harper et al. (16). The C-terminal portion of mouse IκBα (244–314) was inserted into the pAS2 vector. The resultant plasmid pAS2-IκBα-C was transformed into the Y190 yeast strain, and Y190-IκBα-C was established. The RE10 cDNA-bearing pVP16 plasmid was transformed into Y190-IκBα-C and analyzed as described above.
To obtain a cDNA for the complete mouse Ubc9/Hus5 (mE2) coding sequence, an oligo(dT)-primed cDNA library was constructed in pBSSK(−) using cDNAs generated from poly(A)+ RNA from WEHI231, mouse B cell line. The nucleotide sequences were determined by the dideoxyribonucleotide chain-termination procedure using modified T7 DNA polymerase (Sequenase 2.0; United States Biochemical). Databank search was done using blast (21). mE2 or Ubc homologs were aligned against each other using the multiple protein alignment program of Geneworks (IntelliGenetics). The calculated percentage of identical amino acids are shown. Databank accession numbers for each of the Ubc families are as follows: mE2; U82627, Ubc9(h); U31933, Hus5/Ubc3(sp); P40984, Ubc9(sc); X82538, CDC34(h); L22005, Ubc2(h); P23567, Ubc2(sp); P23566, Ubc2(sc); P06104, Ubc7(sc); Q02159, Ubc4(sc); P15731, Ubc5(sc); P15732.
Antibodies and Immunoblot Analysis.
Antibodies against mouse mE2 were prepared by immunizing rabbits with a peptide containing N-terminal 13 residues (SGIALSRLAQERK) and affinity-purified on a column of N-terminal peptide. Anti-IκBα IgG fraction was purchased from Santa Cruz Biotechnology. pCMXmE2, pEGFP-C1 mE2, and pEGFP-N1 mE2 were constructed by ligating the full mE2 coding sequence into pCMX, pEGFP-C1, and pEGFP-N1 (CLONTECH).
293T cells were transfected with one of the plasmids described above. After rinsing with cold PBS, cells were scraped off the plates, centrifuged, and frozen on dry ice. Cytoplasmic and nuclear extracts were prepared as described (22) in the presence of phosphatase inhibitors (50 mM NaF/0.1 mM sodium vanadate/10 mM sodium molybdate/20 mM β-glycerophosphate/10 mM p-nitrophenyl phosphate), and protease inhibitors (0.1 mM phenylmethylsulfonyl fluoride, 21 μg/ml aprotinin, 20 μM benzyloxycarbonyl-Leu-Leu-phenylalanine provided by Signal Pharmaceuticals, San Diego). Cellular or nuclear extracts as indicated were fractionated on 15% SDS/polyacrylamide gel, transferred to Nitropure membranes (Micron Separations, Westboro, MA). Specific proteins were detected by incubation with the appropriate antibody (either the anti-mE2 or anti-IκBα) followed by visualization using ECL luminescence kit (Amersham).
Immunohistochemistry and Histochemistry with Green Fluorescence Protein (GFP).
Cells, either nontransfected or transfected with mE2 expression vector or GFP fusion protein-expressing vector, cultured on coverslips were washed in PBS, and fixed with 1:1 acetone/methanol at −10°C for 1 min and allowed to air dry for 20 min. This was followed by three washes in PBS and a 1-hr block in 2% normal goat serum. The samples were incubated for 1 hr at room temperature with affinity purified rabbit antibodies diluted 1:500 in 2% normal goat serum, then washed three times with cold PBS, and incubated with the secondary antibody [donkey anti-rabbit either fluorescein isothiocyanate or rhodamine (Jackson ImmunoResearch) diluted 1:100]. After rinsing in PBS, the DNA was stained with DAPI (4′,6-diamidino-2-phenylindole; Sigma) made up at 1 μg/ml in PBS, for 5 min, rinsed again, then mounted in a drop of anti-quenching mountant containing 1 mg/ml p-phenylenediamine in 90% glycerol. Preparations were examined to detect immunoreactivities or GFP fluorescence, and photographed on a Nikon Microphot-SA immunofluorescence microscope (23, 24).
Coimmunoprecipitations.
35S-labeled in vitro-translated mE2 and mouse IκBα proteins were incubated with either anti-mE2 antibodies or preimmune sera and protein A-Sepharose beads (Pharmacia) in RIPA buffer (100 mM NaCl/20 mM Tris, pH 8.0/1% Nonidet P-40) for 8 hr at 4°C with constant rotation. The beads were then washed four times with RIPA buffer, followed by boiling in SDS/PAGE loading buffer. The eluted proteins were separated by electrophoresis on a 12% SDS/polyacrylamide gel, visualized by fluorography.
Glutathione S-Transferase (GST)-Pulldown Assay with Reversible Crosslinker.
A negative mutant form of mE2 (mE2*), which, respectively, contains arginine and alanine instead of cysteine-93 and leucine-97, was generated by PCR-based site-directed mutagenesis. pGSTmE2 and pGSTmE2* were constructed by ligation of the full mE2 and mE2* coding sequences, respectively. Plasmids were transformed into Escherichia coli strain BL21, and GST fusion proteins were purified with GST-CL4B (Pharmacia).
GST-pulldown assay was done in the presence of dithiobis-succinimidylpropionate, a reversible crosslinker as follows (25). Five microliters of 35S-labeled in vitro-translated mE2 or mE2* protein was preincubated with 20 μg of GST protein and 5 μl of glutathione-Sepharose 4B beads (Pharmacia) in 100 μl of the crosslinking buffer [100 mM NaCl/20 mM Hepes⋅NaOH, pH 7.9/1 mM DTT/1 mM EDTA/10% glycerol/0.5% Nonidet P-40/1 mM phenylmethylsulfonyl fluoride/1% (vol/vol) aprotinin] for 1 hr at room temperature, and the supernatant was recovered after centrifugation. Five micrograms of GST protein or various GST fusion proteins indicated in the figure legends was added, and the reaction volume was adjusted to 460 μl with crosslinking buffer and incubated for 30 min at room temperature. Forty microliters of 25 mM dithiobissuccinimidylpropionate (Pierce) was added and incubated for 30 min at room temperature. Fifty microliters of 1 M ethanolamine was added and incubated for 15 min at room temperature to quench the reaction. Beads were washed with the crosslinking buffer five times at 4°C and boiled in SDS/PAGE loading buffer. The eluted proteins were separated by electrophoresis on a 12% SDS/polyacrylamide gel and visualized by fluorography, and the band intensities were measured by the PhosphoImager (Molecular Dynamics).
Adenovirus Infection and TNFα Stimulation.
pAxCAmE2 and pAxCAmE2* were constructed by inserting mE2 and mE2* into the replication deficient adenovirus vector, pAxCA, respectively (26, 27). Adenovirus particles were prepared, and HeLa cells were infected at each multiplicity of infection (moi) indicated in the figure legends. Twenty-four hours after infection, cells were treated with recombinant human TNFα (10 ng/ml) (Calbiochem) for the indicated periods. After rinsing with ice-cold PBS, cells were scraped off the plates, centrifuged, and frozen on dry ice. Cytoplasmic and nuclear extracts were prepared as described above.
Electrophoretic Mobility-Shift Assay (EMSA).
EMSA was performed with 5 μg of nuclear extracts and 32P-labeled Igκ κB probe or Oct1 probe as described (22). Products were analyzed on a 4% acrylamide gel, and the band intensities were measured by the PhosphoImager (Molecular Dynamics).
RESULTS
cDNA Cloning of the Mouse Ubc9/Hus5 Homologue.
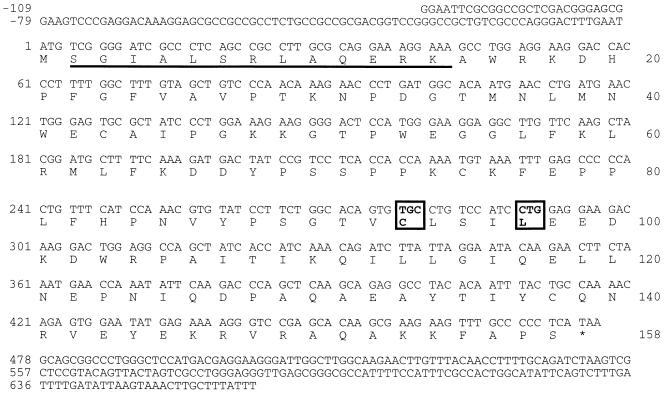
To identify proteins involved in the NF-κB signal transduction pathway, Rel-associated proteins were cloned using the yeast two-hybrid system. The amino(N)-terminal half of mouse cRel was used as a bait to screen a WEHI231 mouse B cell cDNA library. Two overlapping cDNAs were cloned based on the association of their protein product with cRel. The sequence of these cDNAs is homologous to Ubc9 and Hus5, the E2 or Ub-conjugating enzyme family of S. cerevisiae and S. pombe, respectively (28, 29). Additionally, the nucleotide sequence of this mouse Ubc9/Hus5 cDNA (mE2) is identical to the human homologue (30). mE2 has a 158-aa ORF with 66% identity to Hus5 and 56% to Ubc9 (Table 1; Fig. 1). mE2 also shares 30–50% identity with other E2 proteins like Ubc2, Ubc4, and Ubc5 (Table 1). Conserved among mouse, human, and yeast E2 proteins are cysteine-93 and leucine-97. These amino acids are proposed to have a critical role in Ub-conjugation activity (31).
Table 1.
The identity between mE2 and Ubc families
| mE2 | Ubc9 (h) | Hus5/Ubc3 (sp) | Ubc9 (sc) | CDC34 (h) | Ubc2 (h) | Ubc2 (sp) | Ubc2 (sc) | Ubc7 (sc) | Ubc4 (sc) | |
|---|---|---|---|---|---|---|---|---|---|---|
| mE2 | ||||||||||
| Ubc9 (h) | 100 | |||||||||
| Hus5/Ubc3 (sp) | 66 | 66 | ||||||||
| Ubc9 (sc) | 56 | 56 | 61 | |||||||
| CDC34 (h) | 45 | 45 | 21 | 19 | ||||||
| Ubc2 (h) | 41 | 41 | 39 | 37 | 23 | |||||
| Ubc2 (sp) | 41 | 41 | 39 | 35 | 21 | 70 | ||||
| Ubc2 (sc) | 39 | 39 | 35 | 31 | 22 | 61 | 67 | |||
| Ubc7 (sc) | 39 | 39 | 34 | 31 | 27 | 37 | 38 | 33 | ||
| Ubc4 (sc) | 36 | 36 | 34 | 34 | 19 | 36 | 36 | 34 | 35 | |
| Ubc5 (sc) | 35 | 35 | 35 | 36 | 19 | 38 | 37 | 34 | 34 | 93 |
Each of the mE2 or Ubc homologs was aligned against each other using the multiple protein alignment program of Geneworks. The calculated percentage of identical amino acids are shown. See text for databank accession numbers. h, Human; sc, Saccharomyces cerevisiae; sp, Schizosaccharomyces pombe.
Figure 1.
Nucleotide and deduced amino acid sequence of mE2 (mouse homolog of Ubc9/Hus5; GenBank accession no. U82627). The predicted mE2 amino acid sequence is indicated in single-letter code. Cysteine-93 and leucine-97 mutated to generate the dominant-negative mutant mE2* are boxed. The 13 amino acids used to raise the mE2 antisera are underlined.
It is worth pointing out that although mE2 was identified through its association with cRel, it binds more avidly to IκBα.
Cellular Localization of mE2.
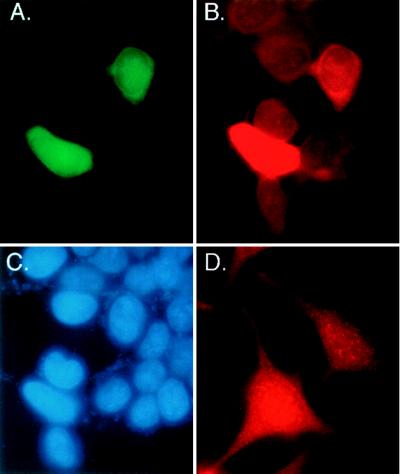
NF-κB activation requires Ub-conjugation and degradation of IκBα, therefore mE2, which associates with the cRel subunit of NF-κB, may function in the Ub-conjugation of IκBα. Because Ub-conjugation of IκBα occurs in the cytoplasm, we tested mE2 protein for cytoplasmic localization. To generate antisera to mE2, we immunized rabbits with a peptide corresponding to the amino-terminal 13 amino acids of mE2 (shown in Fig. 1). Using immunohistochemistry, we examined 293T cells for localization of endogenous mE2 or overexpressed GFP tagged mE2. Fig. 2A shows green fluorescence from GFP-mE2 in both the cytoplasm and the nucleus of 293T cells. Using mE2 antisera, endogenous mE2 also was detected as staining in both the nucleus and cytoplasm of 293T and HeLa cells (Fig. 2 B and D). Similar cellular distribution was observed when C-terminally GFP-tagged mE2 or hemagglutinin-tagged mE2 was expressed from a variety of different promoters (data not shown). In addition to immunohistochemistry, whole-cell extracts from 293T cells were fractionated into cytoplasmic and nuclear fractions and analyzed by Western blot analysis. mE2 antisera detected an 18-kDa protein in both the cytoplasmic and the nuclear fractions (data not shown). We conclude that mE2 is located both in the cytoplasmic and nuclear compartments of the cell.
Figure 2.
Localization of mE2 protein in the cytoplasm and nucleus of mammalian cells. Histochemistry showing endogenous mE2 protein and transiently expressed GFP-mE2 fusion protein localizes in both the cytoplasm and nucleus of human cells. (A) Detection of green fluorescence in both the cytoplasm and nucleus of 293T cells expressing GFP-mE2 fusion protein. (B) mE2-like immunoreactivity was detected in red fluorescence. Two cells expressing GFP-mE2 fusion protein and other cells having endogenous-E2 show the same distribution pattern, both in the cytoplasm and nucleus. (C) The same cells used in A and B were stained with 4′,6-diamidino-2-phenylindole to visualize the nucleus. (D) mE2-like immunoreactivity was detected in red fluorescence in the cytoplasm and nucleus of HeLa cells.
mE2 Binds to IκBα.
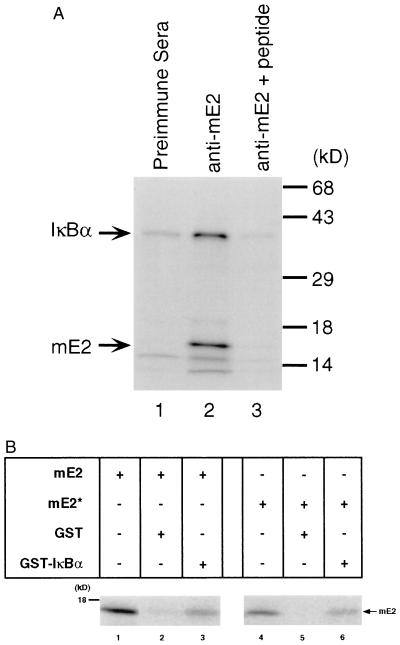
mE2 is found in the cytoplasm and therefore could be involved in the signal-induced degradation of IκBα. To investigate the direct association of mE2 to IκBα, we used both coimmunoprecipitation and GST-pulldown assays. In vitro 35S-labeled mouse IκBα and mE2 were coimmunoprecipitated with mE2 antisera and analyzed by SDS/PAGE. As shown in Fig. 3A, lane 2, IκBα and mE2 can be coimmunoprecipitated by anti-mE2 sera. A very small fraction of a protein (less than 10%) with a molecular weight similar to IκBα was immunoprecipitated by preimmune sera (Fig. 3A, lane 1), which cannot be removed even by preincubation with the peptide used to generate mE2 antisera. Furthermore in Western blot analysis this band comigrating with IκBα cannot be detected (data not shown). No IκBα or mE2 were identified when antisera was preincubated with mE2 peptide (Fig. 3A, lane 3). Additionally, GST-pulldown assays were performed using dithiobissuccinimidylpropionate, a reversible crosslinker. In these assays, 5% of the in vitro-translated mE2 protein associates with GST-IκBα, while less than 0.01% associates with GST alone (Fig. 3B, lanes 2 and 5). The cysteine-93-arginine, leucine-97-alanine mE2 mutant (mE2*) associated with GST-IκBα in a manner similar to wild-type mE2 (Fig. 3B, lane 6). GST-p50, GST-cRel, GST-IκBγ, and GST-Notch ankyrin region also associated with mE2. However, maximum association was observed with GST-IκBα, followed by GST-IκBγ (data not shown). We have extensively investigated the regions of mE2 and IκBα that associate using a large number of deletion mutants in GST-pulldown assays. The overall data suggests that the interaction involves the entire mE2 protein and principally the ankyrin domain of IκBα.
Figure 3.
Specific binding between mE2 and IκBα. (A) 35S-labeled mouse IκBα protein was coimmunoprecipitated with 35S-labeled mE2 protein in vitro by anti-mE2 sera. Equal amounts of protein were loaded in each lane. Coimmunoprecipitation with preimmunesera, anti-mE2 sera, and mE2 peptide competition are shown in lanes 1, 2, and 3, respectively. (B) 35S-labeled mE2 or mE2* protein binding to GST-mIκBα in GST-pulldown assay using dithiobis-succinimidylpropionate, a reversible crosslinker. Ten percent of the in vitro-translated mE2 or mE2* protein was loaded on lanes 1 and 4. Radioactivities were analyzed by PhosphoImager.
We were unable to show association of endogenous or overexpressed mE2 and IκBα in cells. One possible explanation is that mE2 and IκBα only transiently associate during the ubiquitination process. Furthermore the association may be very weak to facilitate the quick on/off rate during the multi-ubiquitination process.
Mutant mE2 Delays IκBα Degradation.
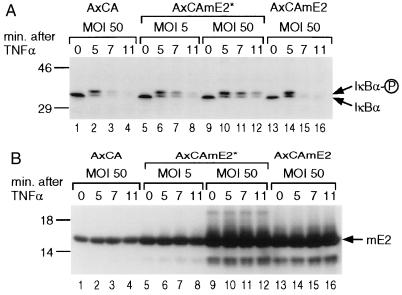
mE2 associates with IκBα in vitro (Fig. 3). To investigate the role of mE2 in signal-induced degradation of IκBα in vivo, we used the transdominant mutant (mE2*) where cysteine-93 and leucine-97 residues in the active site were converted to arginine and alanine (31). To overexpress mE2* in HeLa cells, we used a replication-deficient adenoviral vector. Fig. 4A shows the kinetics of IκBα degradation in the presence and absence of infection with recombinant adenovirus producing the mutant protein, mE2*. Within 5 min of stimulation by TNFα, the IκBα protein undergoes hyperphosphorylation (Fig. 4A, lanes 1, 2 and 13, 14). When cells are infected with vector containing wild-type mE2, the IκBα pool is completely degraded by 7 min (Fig. 4A, lanes 3, 4 and 15, 16). In contrast, IκBα was still present after 11 min in cells infected with vector containing mE2* (Fig. 4A, lanes 7, 8 and 11, 12). When the moi for the mE2*-containing vector was increased from 5 to 50 an even greater proportion of IκBα remains at 11 min poststimulation (Fig. 4A, lanes 9–12). In all cases, hyperphosphorylation of IκBα is unaffected (Fig. 4A, lanes 5–12). As observed by Western blot analysis (Fig. 4B), the amount of mE2* synthesized at moi of 5 is only 2- to 4-fold higher (lanes 5–8) than endogenous mE2. However, at moi of 50 at least 10 times more mE2* is synthesized (Fig. 4B, lanes 9–12). Although the amount of wild-type mE2 overexpression at moi of 50 is comparable to that of the mutant E2 (Fig. 4B, lanes 13–16), the degradation of IκBα is unaffected (Fig. 4A, lanes 13–16). Therefore, we conclude that mE2 is directly involved in degradation of IκBα, and that dominant-negative mE2* inhibits this degradation.
Figure 4.
Kinetics of TNFα-induced degradation of IκBα in cells infected with recombinant adenoviral vectors, (A) HeLa cells were infected with control (empty) (lanes 1–4), mutant mE2* (lanes 5–12), or wild-type mE2 (lanes 13–16) expressing replication-deficient adenovirus vector, AxCA at indicated moi. Twelve hr after infection, 10 ng/ml of rhTNFα was added and cells harvested after indicated times. Twenty-five micrograms of cytoplasmic proteins was blotted and analyzed with IκBα antibody. (B) Protein expression levels were analyzed by Western blotting with anti-mE2 sera.
Dominant-Negative mE2* Delays and Decreases NF-κB Inducible Activity in Vivo.
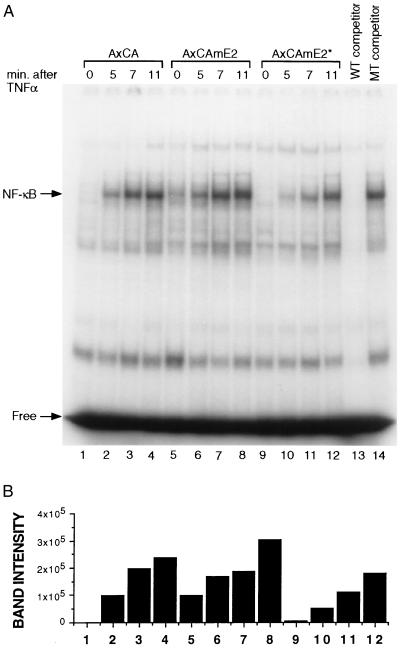
The signal-induced degradation of IκBα leads to the transport of NF-κB proteins to the nucleus and activation of target gene transcription. We therefore reasoned that if the degradation of IκBα is retarded in the presence of mutant E2 protein (mE2*), the kinetics of induction of NF-κB activity should be delayed. NF-κB activity can be measured as an increase in κB-site DNA binding activity in nuclear extracts. Fig. 5A shows DNA binding activity in HeLa cells infected with the vector alone or vector containing wild-type mE2. The kinetics and extent of induction of NF-κB activity are unaffected with respect to uninfected cells (Fig. 5A, lanes 1–8). However, HeLa cells infected with vector containing the mutant E2 showed a significant delay in the kinetics of DNA binding (Fig. 5A, lanes 9 and 10). This delay paralleled the delay of IκBα degradation (Fig. 4A). The DNA binding activity of the unrelated transcription factor Oct1 is not affected by overexpression of either wild-type or mutant mE2 (Fig. 5B). Because the degradation of IκBα is delayed in the presence of mutant mE2 (mE2*), the subsequent activation of DNA binding activity is concomitantly delayed. Thus, it appears that mE2 specifically participates in the degradation of IκBα and thereby leads to NF-κB activation.
Figure 5.
Delayed DNA binding activity. EMSA showing that overexpression of mutant mE2* resulted in a delay in induction of NF-κB DNA binding activity in the HeLa cell nucleus. (A) HeLa cells were infected with control (empty) (lanes 1–4), wild-type mE2 (lanes 5–8), or dominant-negative mutant mE2* (lanes 9–12) expressing adenovirus vector, AxCA at a moi of 5 or 50. TNFα stimulation was done as in Fig. 4A. EMSA was performed with 5 μg of nuclear extract and 32P-labeled NF-κB probe (lanes 1–12). Excess unlabeled wild-type κB competitor was added to the nuclear extract used in lane 13, and mutant κB unlabeled competitor was added to lane 14. (B) The intensity of NF-κB bands shown in A was measured by the PhosphoImager and indicated as bars. Lane numbers are corresponding to that of A. (C) EMSA with Oct1 was performed using the same nuclear extract used in A. Lane numbers correspond to those of A.
DISCUSSION
The activity of NF-κB/Rel family of transcription factors is tightly regulated by its association with inhibitory protein IκB (1, 2). In response to a plethora of external signals, the IκB protein is degraded, allowing the NF-κB proteins to traverse into the nucleus and participate in the transcription of genes containing cognate κB DNA-binding sites (1, 2). The degradation of IκBα, the most well studied member of the IκB family, proceeds in a very orderly fashion. First, the IκBα, which is a phosphoprotein by virtue of phosphorylation at its carboxyl-terminus by casein kinase II, is further phosphorylated at serine residues 32 and 36 in response to the signal (32–34). Although the protein kinase that phosphorylates serine-32 and -36 remains unknown, a high molecular weight complex has been identified by a number of investigators (ref. 11; J. K. Stevenson and I.M.V., unpublished data). After phosphorylation, IκBα, still bound to NF-κB proteins (p50/p65 or p50/Rel complex), is the target of ubiquitination at lysine residues 21 and 22 (9, 10, 35). There also may be additional sites of ubiquitination (lysine-67; K.T., unpublished observations). The multi-ubiquitinated NF-κB/IκB complex then is processed by 26S ATP-dependent proteasome. IκBα is degraded, thereby releasing the NF-κB complex to be transported into the nucleus. The precise mechanism by which NF-κB/IκB complex is processed through the proteasome is not understood. However, the lack of detectable free ubiquitinated IκBα indicates that ubiquitinated IκBα is degraded while still associated with NF-κB (7, 22, 31, 33, 36). In a similar manner p105 is processed to p50 after C-terminal phosphorylation, ubiquitination, and degradation by proteasomes. Similar mechanisms must prevent NF-κB proteins from further proteolytic degradation while allowing degradation or processing of associated IκB or NF-κB precursor proteins (35).
The identification of mE2 as a homologue of Ubc9/Hus5 and its association with IκBα was initially puzzling, because the yeast proteins are localized in the nucleus (28, 29, 37), whereas IκBα is predominantly a cytoplasmic protein. Additionally, the ubiquitinated forms of IκBα are cytoplasmic (1, 2). It was, therefore, gratifying to establish that mE2 was both cytoplasmic and nuclear in its localization (Fig. 2). In addition to mE2, it has been suggested that yeast Ubc4 and Ubc5 can catalyze Ub conjugation on mammalian IκBα (11). E2-F1, a mammalian member of the E2 family, also has been shown to be involved in Ub conjugation of p105 in vitro (37). Our data does not rule out a role for Ubc4/Ubc5 and E2-F1 in the Ub conjugation and degradation of IκBα. Clearly, there are multiple E2 proteins and more than one may have a role in the Ub-conjugation of IκBα (11, 37).
Does mE2 require an E3 or does it directly interact with the substrate IκBα? Although we lack evidence for an IκBα-associated E3 protein, it appears that association of mE2 to IκBα is increased by cellular extracts (M.P. and K.T., unpublished data). Consequently, these extracts may provide an E3 ligase. A novel E3 ligase proposed by Orian et al. (37) may be involved. We currently are identifying proteins that directly interact with mE2 in an effort to identify E3 and other proteins involved in IκBα degradation.
E2 proteins previously have been postulated to be involved in the Ub-conjugated degradation of IκBα, based on in vitro assays (4–12). We provide the first in vivo proof for E2 involvement by using a transdominant-negative mutant of E2. The mutant E2 does not allow Ub ligation to the substrate and consequently interferes with the process of degradation of IκBα. Our data strongly support the role of mE2 in IκBα degradation because in the presence of overexpressed mutant mE2, the kinetics of degradation of IκBα are substantially delayed. Additionally, mutant mE2 somewhat decreases multi-ubiquitination of IκBα in vitro (K.T., unpublished observations). Because the amount of mE2 is substantial in the cell, to overcome endogenous E2 we generated large amounts of the mutant protein using recombinant adenovirus expression vectors. This strategy also allowed us to compare dosage dependence of mutant mE2 expression (5 moi and 50 moi). The adenovirus itself had no effect on the rate of IκBα degradation or NF-κB activation, nor did the overproduction of wild-type mE2 protein (Fig. 4 A and B). Only when the mutant protein was overexpressed was the degradation retarded. The delayed degradation of IκBα directly affects the amount as well as the kinetics of NF-κB activation (Fig. 5). The DNA binding activity of transcription factor Oct1 was not affected (Fig. 5C), indicating that overproduction of either mE2 or mE2* protein does not have general inhibitory effects on other cellular proteins. The mutant mE2 did not delay the degradation of p105, suggesting that mE2 plays a specific role in degradation of IκBα (K.T., unpublished results). What are the consequences of inhibition of IκBα degradation? In another study we have shown that cells producing stable dominant-negative IκBα protein (IκBαM) undergo apoptosis upon induction with TNFα (38). Whether cells infected with adenovirus containing mutant mE2* protein also undergo apoptosis in response to TNFα remains an interesting and open question.
Although there are multiple members of the IκB family, they all do not undergo signal-induced rapid degradation typified by IκBα. For instance, IκBβ is also phosphorylated at comparable serine residues (serine-19 and -23) (39), and is ubiquitinated, but displays significantly slower kinetics of degradation. Bcl3 protein is quite stable, and it is not clear if it undergoes Ub-conjugated degradation. IκBγ most likely undergoes Ub-conjugated degradation, similar to processing of p105 (7, 37). It is not known if each of the IκB proteins undergo signal-induced degradation by Ub-conjugation pathway, or if they use the same or different E2 proteins. The availability of recombinant adenoviruses generating mutant mE2* proteins will be a very helpful tool to decipher the role of Ub-conjugated degradation pathway of IκB proteins.
Acknowledgments
We are indebted to Dr. Jennifer Stevenson for critical reading of the manuscript. We thank Drs. Stephen Elledge, Stan Hollenberg, and Hideo Toyoshima for their kind gifts of pAS2, pVP16, and pAS2-CDK4, respectively. We thank Ezra Wiater, Makoto Kawai, and Daisuke Yabe for technical assistance at various times. We thank Pat McClintock and Mari Tashiro for their help in the preparation of the manuscript. K.T. is supported by a Japan Foundation for Aging and Health Fellowship. M.P. is a graduate student in the Department of Biology at the University of California, San Diego and is supported by the San Diego Fellowship. This work was supported by funds from the National Institutes of Health and the Frances C. Berger Foundation. I.M.V. is an American Cancer Society Professor of Molecular Biology.
ABBREVIATIONS
- mE2
mouse ubiquitin-conjugating enzyme
- TNF
tumor necrosis factor
- Ub
ubiquitin
- Ubc
Ub-conjugating enzyme
- GST
glutathione S-transferase
- GFP
green fluorescence protein
- moi
multiplicity of infection
- EMSA
electrophoretic mobility-shift assay
References
- 1.Verma I M, Stevenson J K, Schwarz E M, Van Antwerp D, Miyamoto S. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 2.Baeuerle P A, Baltimore D. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 3.Beg A A, Sha W C, Bronson R T, Baltimore D. Genes Dev. 1995;9:2736–2746. doi: 10.1101/gad.9.22.2736. [DOI] [PubMed] [Google Scholar]
- 4.Alkalay I, Yaron A, Hatzubai A, Jung S, Avraham A, Gerlitz O, Pashut-Lavon I, Ben-Neriah Y. Mol Cell Biol. 1995;15:1294–1301. doi: 10.1128/mcb.15.3.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alkalay I, Yaron A, Hatzubai A, Orian A, Ciechanover A, Ben-Neriah Y. Proc Natl Acad Sci USA. 1995;92:10599–10603. doi: 10.1073/pnas.92.23.10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z, Hagler J, Palombella V J, Melandri F, Scherer D, Ballard D, Maniatis T. Genes Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 7.Palombella V J, Rando O J, Goldberg A L, Maniatis T. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez M S, Michalopoulos I, Arenzana-Seisdedos F, Hay R T. Mol Cell Biol. 1995;15:2413–2419. doi: 10.1128/mcb.15.5.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scherer D C, Brockman J A, Chen Z, Maniatis T, Ballard D W. Proc Natl Acad Sci USA. 1995;92:11259–11263. doi: 10.1073/pnas.92.24.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baldi L, Brown K, Franzoso G, Siebenlist U. J Biol Chem. 1996;271:376–379. doi: 10.1074/jbc.271.1.376. [DOI] [PubMed] [Google Scholar]
- 11.Chen Z J, Parent L, Maniatis T. Cell. 1996;84:853–862. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- 12.Roff M, Thompson J, Rodriguez M S, Jacque J M, Baleux F, Arenzana-Seisdedos F, Hay R T. J Biol Chem. 1996;271:7844–7850. doi: 10.1074/jbc.271.13.7844. [DOI] [PubMed] [Google Scholar]
- 13.Finco T S, Baldwin A S. Immunity. 1995;3:263–272. doi: 10.1016/1074-7613(95)90112-4. [DOI] [PubMed] [Google Scholar]
- 14.Ciechanover A. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 15.Chau V, Tobias J W, Bachmair A, Marriott D, Ecker D J, Gonda D K, Varshavsky A. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 16.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 17.Durfee T, Becherer K, Chen P L, Yeh S H, Yang Y A, Kilburn E, Lee W H, Elledge S J. Genes Dev. 1993;7:555–556. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 18.Toyoshima H, Hunter T. Cell. 1994;78:67–74. doi: 10.1016/0092-8674(94)90573-8. [DOI] [PubMed] [Google Scholar]
- 19.Vojtek A B, Hollenberg S M, Cooper J A. Cell. 1993;74:205–221. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 20.Tashiro K, Nakano T, Honjo T. Methods Mol Biol. 1996;69:203–219. doi: 10.1385/0-89603-383-x:203. [DOI] [PubMed] [Google Scholar]
- 21.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Miyamoto S, Maki M, Schmitt M J, Hatanaka M, Verma I M. Proc Natl Acad Sci USA. 1994;91:12740–12744. doi: 10.1073/pnas.91.26.12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dyck J A, Maul G G, Miller W, Jr, Chen J D, Kakizuka A, Evans R M. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 24.Ogawa H, Inouye S, Tsuji F I, Yasuda K, Umesono K. Proc Natl Acad Sci USA. 1995;92:11899–11903. doi: 10.1073/pnas.92.25.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ransone L J. Methods Enzymol. 1995;254:491–497. doi: 10.1016/0076-6879(95)54034-2. [DOI] [PubMed] [Google Scholar]
- 26.Hashimoto M, Aruga J, Hosoya Y, Kanegae Y, Saito I, Mikoshiba K. Hum Gene Ther. 1996;7:149–158. doi: 10.1089/hum.1996.7.2-149. [DOI] [PubMed] [Google Scholar]
- 27.Miyake S, Makimura M, Kanegae Y, Harada S, Sato Y, Takamori K, Tokuda C, Saito I. Proc Natl Acad Sci USA. 1996;93:1320–1324. doi: 10.1073/pnas.93.3.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Al-Khodairy F, Enoch T, Hagan I M, Carr A M. J Cell Sci. 1995;108:475–486. doi: 10.1242/jcs.108.2.475. [DOI] [PubMed] [Google Scholar]
- 29.Seufert W, Futcher B, Jentsch S. Nature (London) 1995;373:78–81. doi: 10.1038/373078a0. [DOI] [PubMed] [Google Scholar]
- 30.Kovalenko O V, Plug A W, Haaf T, Gonda D K, Ashley T, Ward D C, Radding C M, Golub E I. Proc Natl Acad Sci USA. 1996;93:2958–2963. doi: 10.1073/pnas.93.7.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banerjee A, Deshaies R J, Chau V. J Biol Chem. 1995;270:26209–26215. doi: 10.1074/jbc.270.44.26209. [DOI] [PubMed] [Google Scholar]
- 32.Barroga C F, Stevenson J K, Schwarz M E, Verma I M. Proc Natl Acad Sci USA. 1995;92:7637–7641. doi: 10.1073/pnas.92.17.7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown K, Gerstberger S, Carlson L, Franzoso G, Siebenlist U. Science. 1995;267:1485–1488. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- 34.Schwarz E M, Van Antwerp D, Verma I M. Mol Cell Biol. 1996;16:3554–3559. doi: 10.1128/mcb.16.7.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DiDonato J, Mercurio F, Rosette C, Wu-Li J, Suyang H, Ghosh S, Karin M. Mol Cell Biol. 1996;16:1295–1304. doi: 10.1128/mcb.16.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Traenckner E B-M, Wilk S, Baeuerle P A. EMBO J. 1994;13:5433–5441. doi: 10.1002/j.1460-2075.1994.tb06878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orian A, Whiteside S, Israel A, Stancovski I, Schwartz A L, Ciechanover A. J Biol Chem. 1995;270:21707–21714. doi: 10.1074/jbc.270.37.21707. [DOI] [PubMed] [Google Scholar]
- 38.Van Antwerp D J, Martin S J, Kafri T, Green D R, Verma I M. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 39.Thompson J E, Phillips R J, Erdjument-Bromage H, Tempst P, Ghosh S. Cell. 1995;80:573–582. doi: 10.1016/0092-8674(95)90511-1. [DOI] [PubMed] [Google Scholar]