Abstract
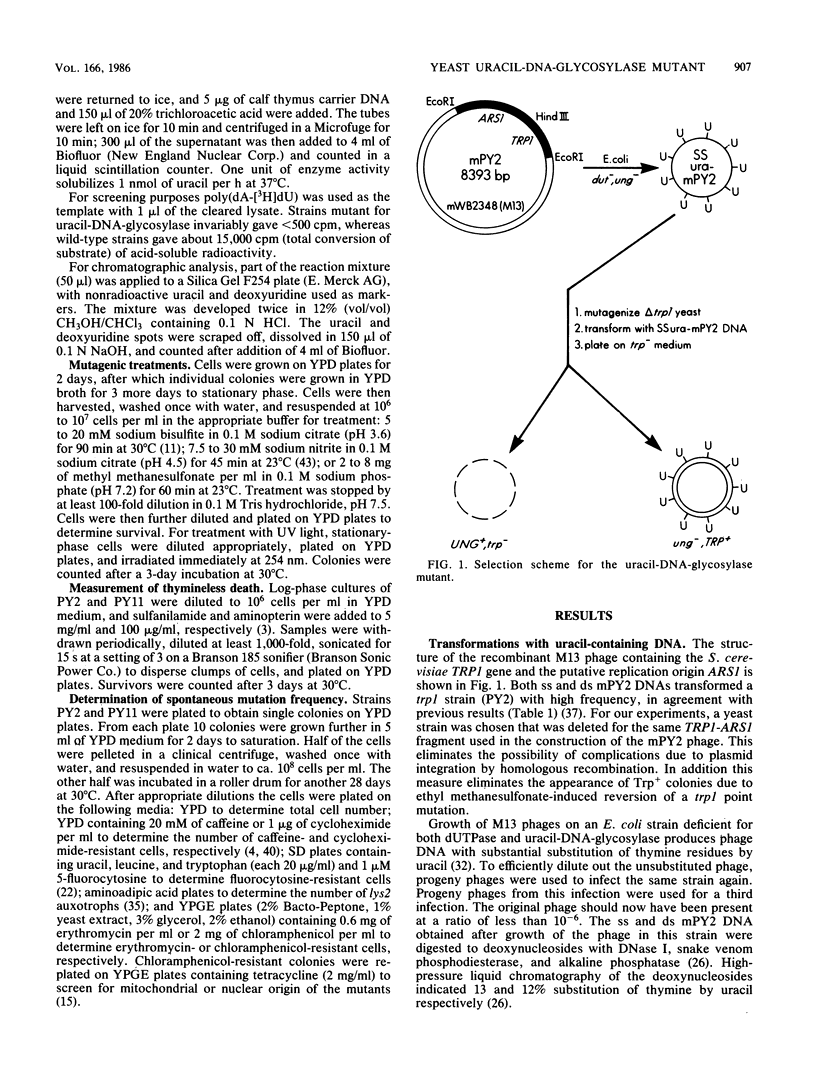
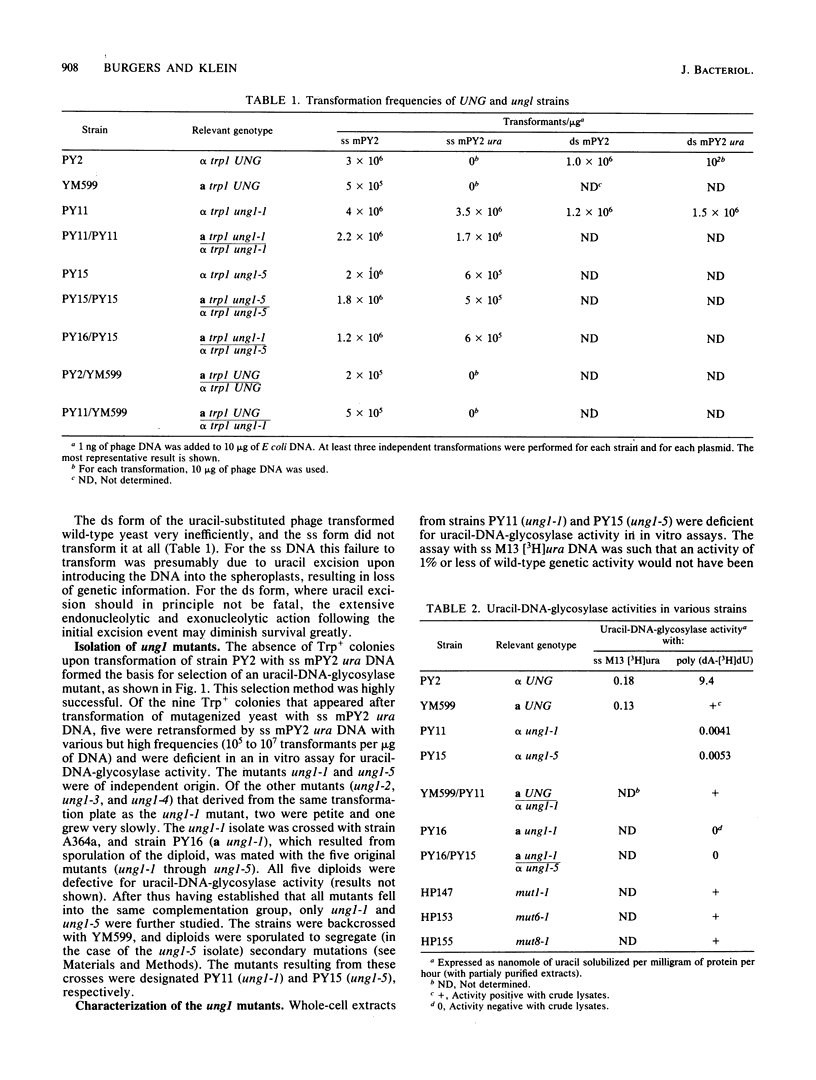
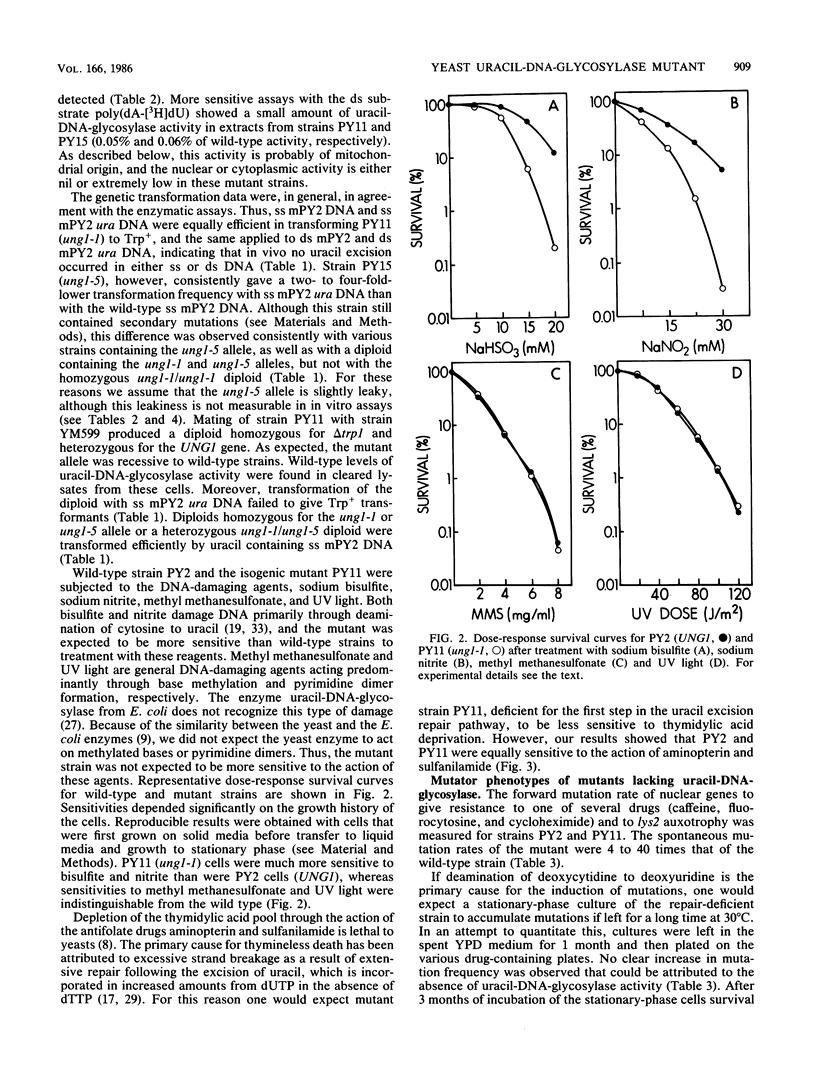
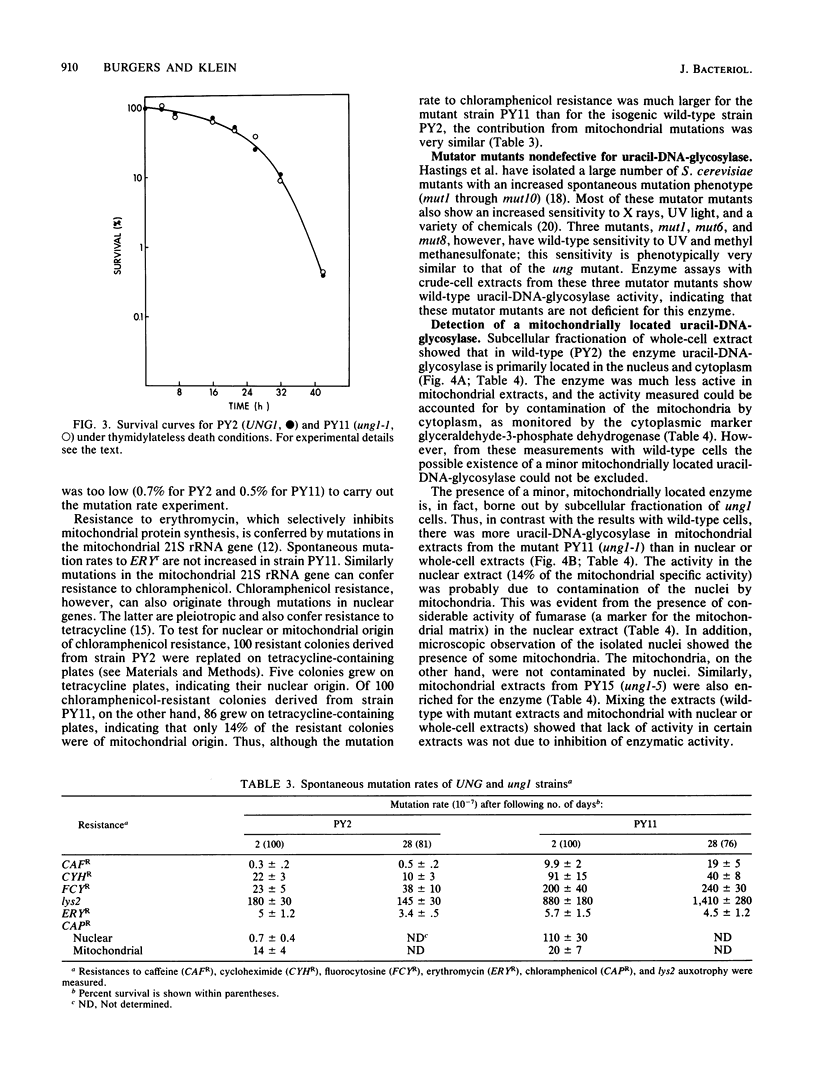
A coliphage M13 chimer containing the Saccharomyces cerevisiae TRP1 gene and ARS1 replication origin (mPY2) was grown on an ung- dut- strain of Escherichia coli. The resulting single-stranded phage DNA had 13% of thymine residues substituted by uracil. This DNA failed to transform a delta trp1 yeast strain to prototrophy. However, when a mutagenized yeast stock was transformed with uracil-containing single-stranded mPY2 DNA, unstable transformants were obtained. After plasmid segregation, about half of these were retransformed at a high frequency by uracil-containing single-stranded mPY2 DNA. In vitro, these mutants were defective for uracil-DNA-glycosylase activity. They were designated ung1. Strains containing the ung1 mutation have an increased sensitivity to sodium bisulfite and sodium nitrite but a wild-type sensitivity to methyl methanesulfonate, UV light, and drugs that cause depletion of the thymidylate pool. They have a moderate mutator phenotype for nuclear but not for mitochondrial genes. A low mitochondrial uracil-DNA-glycosylase activity was demonstrated in the mutant strains.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson C. T., Friedberg E. C. The presence of nuclear and mitochondrial uracil-DNA glycosylase in extracts of human KB cells. Nucleic Acids Res. 1980 Feb 25;8(4):875–888. [PMC free article] [PubMed] [Google Scholar]
- Barclay B. J., Kunz B. A., Little J. G., Haynes R. H. Genetic and biochemical consequences of thymidylate stress. Can J Biochem. 1982 Mar;60(3):172–184. doi: 10.1139/o82-023. [DOI] [PubMed] [Google Scholar]
- Barclay B. J., Little J. G. Genetic damage during thymidylate starvation in Saccharomyces cerevisiae. Mol Gen Genet. 1978 Mar 20;160(1):33–40. doi: 10.1007/BF00275116. [DOI] [PubMed] [Google Scholar]
- Barnes W. M., Bevan M. Kilo-sequencing: an ordered strategy for rapid DNA sequence data acquisition. Nucleic Acids Res. 1983 Jan 25;11(2):349–368. doi: 10.1093/nar/11.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brendel M., Langjahr U. G. "Thymineless death" in a strain of Saccharomyces cerevisiae auxotrophic for deoxythymidine-5'-monophosphate. Mol Gen Genet. 1974;131(4):351–358. doi: 10.1007/BF00264865. [DOI] [PubMed] [Google Scholar]
- Crosby B., Prakash L., Davis H., Hinkle D. C. Purification and characterization of a uracil-DNA glycosylase from the yeast. Saccharomyces cerevisiae. Nucleic Acids Res. 1981 Nov 11;9(21):5797–5809. doi: 10.1093/nar/9.21.5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Roza R., Friedberg E. C., Duncan B. K., Warner H. R. Repair of nitrous acid damage to DNA in Escherichia coli. Biochemistry. 1977 Nov 1;16(22):4934–4939. doi: 10.1021/bi00641a030. [DOI] [PubMed] [Google Scholar]
- Dorange J. L., Dupuy P. Mise en évidence d'une action mutagène du sulfite de sodium sur la levure. C R Acad Sci Hebd Seances Acad Sci D. 1972 May 15;274(20):2798–2800. [PubMed] [Google Scholar]
- Duncan B. K., Rockstroh P. A., Warner H. R. Escherichia coli K-12 mutants deficient in uracil-DNA glycosylase. J Bacteriol. 1978 Jun;134(3):1039–1045. doi: 10.1128/jb.134.3.1039-1045.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan B. K., Weiss B. Specific mutator effects of ung (uracil-DNA glycosylase) mutations in Escherichia coli. J Bacteriol. 1982 Aug;151(2):750–755. doi: 10.1128/jb.151.2.750-755.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster J. L., Kleese R. A. The segregation of mitochondrial genes in yeast. II. Analysis of zygote pedigrees of drug-resistant X drug-sensitive crosses. Mol Gen Genet. 1975 Sep 8;139(4):341–355. doi: 10.1007/BF00267974. [DOI] [PubMed] [Google Scholar]
- Goulian M., Bleile B., Tseng B. Y. Methotrexate-induced misincorporation of uracil into DNA. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1956–1960. doi: 10.1073/pnas.77.4.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings P. J., Quah S. K., von Borstel R. C. Spontaneous mutation by mutagenic repair of spontaneous lesions in DNA. Nature. 1976 Dec 23;264(5588):719–722. doi: 10.1038/264719a0. [DOI] [PubMed] [Google Scholar]
- Hayatsu H. Bisulfite modification of nucleic acids and their constituents. Prog Nucleic Acid Res Mol Biol. 1976;16:75–124. doi: 10.1016/s0079-6603(08)60756-4. [DOI] [PubMed] [Google Scholar]
- Jensen R., Sprague G. F., Jr, Herskowitz I. Regulation of yeast mating-type interconversion: feedback control of HO gene expression by the mating-type locus. Proc Natl Acad Sci U S A. 1983 May;80(10):3035–3039. doi: 10.1073/pnas.80.10.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jund R., Lacroute F. Genetic and physiological aspects of resistance to 5-fluoropyrimidines in Saccharomyces cerevisiae. J Bacteriol. 1970 Jun;102(3):607–615. doi: 10.1128/jb.102.3.607-615.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo K. C., McCune R. A., Gehrke C. W., Midgett R., Ehrlich M. Quantitative reversed-phase high performance liquid chromatographic determination of major and modified deoxyribonucleosides in DNA. Nucleic Acids Res. 1980 Oct 24;8(20):4763–4776. doi: 10.1093/nar/8.20.4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T., Ljungquist S., Siegert W., Nyberg B., Sperens B. DNA N-glycosidases: properties of uracil-DNA glycosidase from Escherichia coli. J Biol Chem. 1977 May 25;252(10):3286–3294. [PubMed] [Google Scholar]
- Lindahl T., Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974 Jul 30;13(16):3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- Makino F., Munakata N. Deoxyuridine residues in DNA of thymine-requiring Bacillus subtilis strains with defective N-glycosidase activity for uracil-containing DNA. J Bacteriol. 1978 Apr;134(1):24–29. doi: 10.1128/jb.134.1.24-29.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera B. M. DNA intermediates at the Escherichia coli replication fork: effect of dUTP. Proc Natl Acad Sci U S A. 1978 Jan;75(1):238–242. doi: 10.1073/pnas.75.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RACKER E. Spectrophotometric measurements of the enzymatic formation of fumaric and cis-aconitic acids. Biochim Biophys Acta. 1950 Jan;4(1-3):211–214. doi: 10.1016/0006-3002(50)90026-6. [DOI] [PubMed] [Google Scholar]
- Sagher D., Strauss B. Insertion of nucleotides opposite apurinic/apyrimidinic sites in deoxyribonucleic acid during in vitro synthesis: uniqueness of adenine nucleotides. Biochemistry. 1983 Sep 13;22(19):4518–4526. doi: 10.1021/bi00288a026. [DOI] [PubMed] [Google Scholar]
- Scott J. H., Schekman R. Lyticase: endoglucanase and protease activities that act together in yeast cell lysis. J Bacteriol. 1980 May;142(2):414–423. doi: 10.1128/jb.142.2.414-423.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons R. R., Friedberg E. C. Enzymatic degradation of uracil-containing deoxyribonucleic acid. V. Survival of Escherichia coli and coliphages treated with sodium bisulfite. J Bacteriol. 1979 Mar;137(3):1243–1252. doi: 10.1128/jb.137.3.1243-1252.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H., Bieker J. J., Dumas L. B. Genetic transformation of Saccharomyces cerevisiae with single-stranded circular DNA vectors. Gene. 1982 Dec;20(3):441–449. doi: 10.1016/0378-1119(82)90213-x. [DOI] [PubMed] [Google Scholar]
- Stinchcomb D. T., Struhl K., Davis R. W. Isolation and characterisation of a yeast chromosomal replicator. Nature. 1979 Nov 1;282(5734):39–43. doi: 10.1038/282039a0. [DOI] [PubMed] [Google Scholar]
- Tye B. K., Chien J., Lehman I. R., Duncan B. K., Warner H. R. Uracil incorporation: a source of pulse-labeled DNA fragments in the replication of the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1978 Jan;75(1):233–237. doi: 10.1073/pnas.75.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILKIE D., LEE B. K. GENETIC ANALYSIS OF ACTIDIONE RESISTANCE IN SACCHAROMYCES CEREVISIAE. Genet Res. 1965 Feb;6:130–138. doi: 10.1017/s0016672300003992. [DOI] [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]



