Abstract
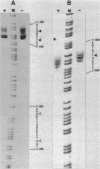
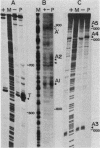
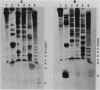
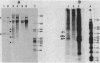
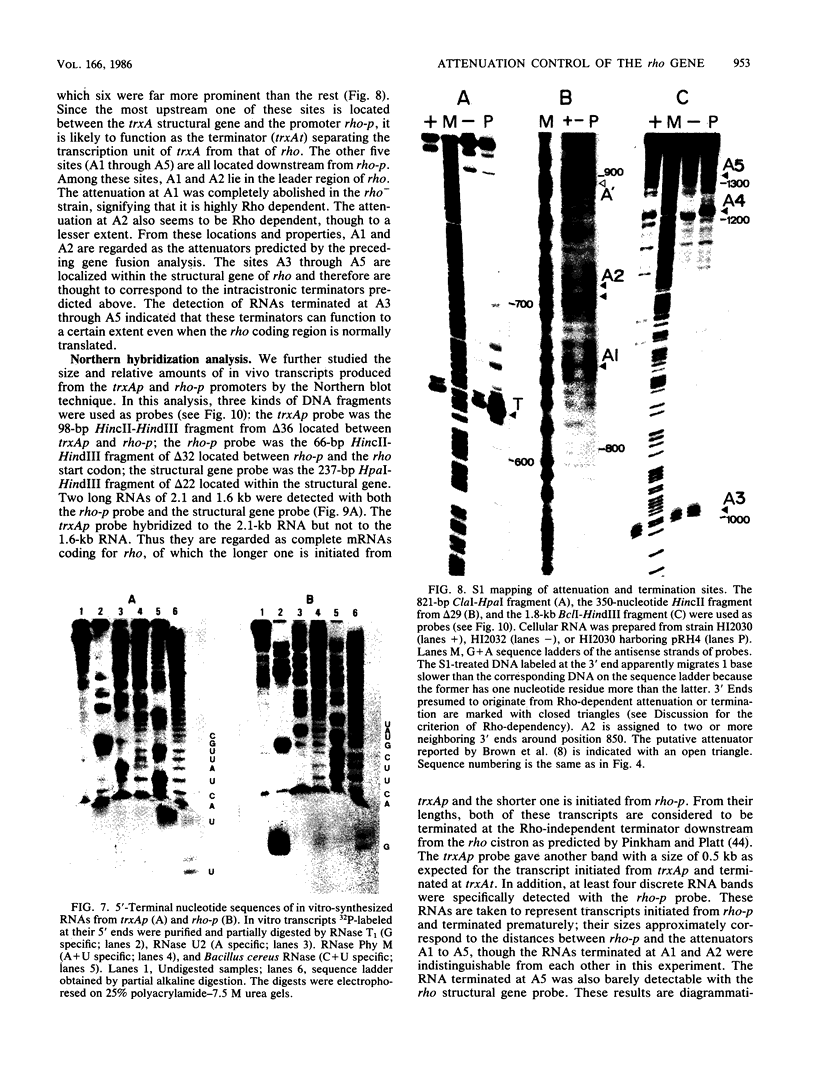
We present evidence that the expression of rho is regulated by rho-dependent attenuation of transcription. Gene fusion analysis with nested series of deletions of rho indicated that the transcription of rho is attenuated in a rho-dependent manner in the leader region and that neither a read-through transcription from the upstream gene, trxA, nor a modulation of transcription initiation of the rho promoter is involved in the self-control of rho. S1 mapping and Northern hybridization analyses localized at least six transcription attenuation or termination sites in the region ranging from the 3' end of the trxA structural gene to the middle of the rho structural gene. Among them, the most upstream site overlapping the rho promoter sequence was assigned to the terminator for the trxA gene, and the second and third sites, mapping about 80 and 50 nucleotides upstream from the start codon of rho, were suggested to function as the major attenuation sites for regulation of the rho expression. Further, the start points of the trxA and rho RNAs were determined in an in vitro transcription system to be located 111 nucleotides (U) and 255 nucleotides (G) upstream from their respective start codons. These results necessitate revisions of previous predictions on the sites of transcriptional signals in the trxA and rho genes (S. Brown, B. Albrechtsen, S. Pedersen, and P. Klemm, J. Mol. Biol. 162:283-298, 1982; C.-J. Lim, D. Geraghty, and J. A. Fuchs, J. Bacteriol. 163:311-316, 1985; B.J. Wallace and S.R. Kushner, Gene 32:399-408, 1984).
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Gottesman M. Control of transcription termination. Annu Rev Biochem. 1978;47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- Aiba H., Adhya S., de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981 Nov 25;256(22):11905–11910. [PubMed] [Google Scholar]
- Aksoy S., Squires C. L., Squires C. Translational coupling of the trpB and trpA genes in the Escherichia coli tryptophan operon. J Bacteriol. 1984 Feb;157(2):363–367. doi: 10.1128/jb.157.2.363-367.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barik S., Bhattacharya P., Das A. Autogenous regulation of transcription termination factor Rho. J Mol Biol. 1985 Apr 20;182(4):495–508. doi: 10.1016/0022-2836(85)90236-0. [DOI] [PubMed] [Google Scholar]
- Blumenthal R. M., Reeh S., Pedersen S. Regulation of transcription factor rho and the alpha subunit of RNA polymerase in Escherichia coli B/r. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2285–2288. doi: 10.1073/pnas.73.7.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Brown S., Albrechtsen B., Pedersen S., Klemm P. Localization and regulation of the structural gene for transcription-termination factor rho of Escherichia coli. J Mol Biol. 1982 Dec 5;162(2):283–298. doi: 10.1016/0022-2836(82)90527-7. [DOI] [PubMed] [Google Scholar]
- Burgess R. R., Jendrisak J. J. A procedure for the rapid, large-scall purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry. 1975 Oct 21;14(21):4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Christiansen L., Pedersen S. Cloning, restriction endonuclease mapping and post-transcriptional regulation of rpsA, the structural gene for ribosomal protein S1. Mol Gen Genet. 1981;181(4):548–551. doi: 10.1007/BF00428751. [DOI] [PubMed] [Google Scholar]
- Das A., Court D., Adhya S. Isolation and characterization of conditional lethal mutants of Escherichia coli defective in transcription termination factor rho. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1959–1963. doi: 10.1073/pnas.73.6.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Wolska K. Transcription antitermination in vitro by lambda N gene product: requirement for a phage nut site and the products of host nusA, nusB, and nusE genes. Cell. 1984 Aug;38(1):165–173. doi: 10.1016/0092-8674(84)90537-3. [DOI] [PubMed] [Google Scholar]
- Das A., Yanofsky C. A ribosome binding site sequence is necessary for efficient expression of the distal gene of a translationally-coupled gene pair. Nucleic Acids Res. 1984 Jun 11;12(11):4757–4768. doi: 10.1093/nar/12.11.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H. Phy M: an RNase activity specific for U and A residues useful in RNA sequence analysis. Nucleic Acids Res. 1980 Jul 25;8(14):3133–3142. doi: 10.1093/nar/8.14.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebright R. H., Cossart P., Gicquel-Sanzey B., Beckwith J. Mutations that alter the DNA sequence specificity of the catabolite gene activator protein of E. coli. Nature. 1984 Sep 20;311(5983):232–235. doi: 10.1038/311232a0. [DOI] [PubMed] [Google Scholar]
- Enger-Valk B. E., van Rotterdam J., Pouwels P. H. The construction of new vehicles for the cloning of transcription termination signals. Nucleic Acids Res. 1981 Apr 24;9(8):1973–1989. doi: 10.1093/nar/9.8.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnham P. J., Greenblatt J., Platt T. Effects of NusA protein on transcription termination in the tryptophan operon of Escherichia coli. Cell. 1982 Jul;29(3):945–951. doi: 10.1016/0092-8674(82)90457-3. [DOI] [PubMed] [Google Scholar]
- Friedman D. I., Schauer A. T., Baumann M. R., Baron L. S., Adhya S. L. Evidence that ribosomal protein S10 participates in control of transcription termination. Proc Natl Acad Sci U S A. 1981 Feb;78(2):1115–1118. doi: 10.1073/pnas.78.2.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischauf A. M., Garoff H., Lehrach H. A subcloning strategy for DNA sequence analysis. Nucleic Acids Res. 1980 Dec 11;8(23):5541–5549. doi: 10.1093/nar/8.23.5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh B., Das A. nusB: a protein factor necessary for transcription antitermination in vitro by phage lambda N gene product. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6305–6309. doi: 10.1073/pnas.81.20.6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt J., McLimont M., Hanly S. Termination of transcription by nusA gene protein of Escherichia coli. Nature. 1981 Jul 16;292(5820):215–220. doi: 10.1038/292215a0. [DOI] [PubMed] [Google Scholar]
- Imai M., Shigesada K. Studies on the altered rho factor in a nitA mutants of Escherichia coli defective in transcription termination. I. Characterization and quantitative determination of rho in cell extracts. J Mol Biol. 1978 Apr 25;120(4):451–466. doi: 10.1016/0022-2836(78)90348-0. [DOI] [PubMed] [Google Scholar]
- Inoko H., Imai M. Isolation and genetic characterization of the nitA mutants of Escherichia coli affecting the termination factor rho. Mol Gen Genet. 1976 Jan 16;143(2):211–221. doi: 10.1007/BF00266924. [DOI] [PubMed] [Google Scholar]
- Inoko H., Shigesada K., Imai M. Isolation and characterization of conditional-lethal rho mutants of Escherichia coli. Proc Natl Acad Sci U S A. 1977 Mar;74(3):1162–1166. doi: 10.1073/pnas.74.3.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston R. E., Chamberlin M. J. Pausing and attenuation of in vitro transcription in the rrnB operon of E. coli. Cell. 1981 Dec;27(3 Pt 2):523–531. doi: 10.1016/0092-8674(81)90394-9. [DOI] [PubMed] [Google Scholar]
- Korn L. J., Yanofsky C. Polarity suppressors defective in transcription termination at the attenuator of the tryptophan operon of Escherichia coli have altered rho factor. J Mol Biol. 1976 Sep 15;106(2):231–241. doi: 10.1016/0022-2836(76)90082-6. [DOI] [PubMed] [Google Scholar]
- Kung H., Bekesi E., Guterman S. K., Gray J. E., Traub L., Calhoun D. H. Autoregulation of the rho gene of Escherichia coli K-12. Mol Gen Genet. 1984;193(2):210–213. doi: 10.1007/BF00330669. [DOI] [PubMed] [Google Scholar]
- Küpper H., Sekiya T., Rosenberg M., Egan J., Landy A. A rho-dependent termination site in the gene coding for tyrosine tRNA su3 of Escherichia coli. Nature. 1978 Mar 30;272(5652):423–428. doi: 10.1038/272423a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lau L. F., Roberts J. W., Wu R. Transcription terminates at lambda tR1 in three clusters. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6171–6175. doi: 10.1073/pnas.79.20.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C. J., Geraghty D., Fuchs J. A. Cloning and nucleotide sequence of the trxA gene of Escherichia coli K-12. J Bacteriol. 1985 Jul;163(1):311–316. doi: 10.1128/jb.163.1.311-316.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockard R. E., Alzner-Deweerd B., Heckman J. E., MacGee J., Tabor M. W., RajBhandary U. L. Sequence analysis of 5'[32P] labeled mRNA and tRNA using polyacrylamide gel electrophoresis. Nucleic Acids Res. 1978 Jan;5(1):37–56. doi: 10.1093/nar/5.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G., Morris N. R. Biological function for 6-methyladenine residues in the DNA of Escherichia coli K12. J Mol Biol. 1974 May 15;85(2):309–322. doi: 10.1016/0022-2836(74)90366-0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Morgan W. D., Bear D. G., Litchman B. L., von Hippel P. H. RNA sequence and secondary structure requirements for rho-dependent transcription termination. Nucleic Acids Res. 1985 May 24;13(10):3739–3754. doi: 10.1093/nar/13.10.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott J. E., Grant R. A., Ho Y. S., Platt T. Maximizing gene expression from plasmid vectors containing the lambda PL promoter: strategies for overproducing transcription termination factor rho. Proc Natl Acad Sci U S A. 1985 Jan;82(1):88–92. doi: 10.1073/pnas.82.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkham J. L., Platt T. The nucleotide sequence of the rho gene of E. coli K-12. Nucleic Acids Res. 1983 Jun 11;11(11):3531–3545. doi: 10.1093/nar/11.11.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner D. Evidence that mutations in the suA polarity suppressing gene directly affect termination factor rho. Nature. 1976 Jan 15;259(5539):151–153. doi: 10.1038/259151a0. [DOI] [PubMed] [Google Scholar]
- Richardson J. P., Grimley C., Lowery C. Transcription termination factor rho activity is altered in Escherichia coli with suA gene mutations. Proc Natl Acad Sci U S A. 1975 May;72(5):1725–1728. doi: 10.1073/pnas.72.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J. W. Termination factor for RNA synthesis. Nature. 1969 Dec 20;224(5225):1168–1174. doi: 10.1038/2241168a0. [DOI] [PubMed] [Google Scholar]
- Rubin C. M., Schmid C. W. Pyrimidine-specific chemical reactions useful for DNA sequencing. Nucleic Acids Res. 1980 Oct 24;8(20):4613–4619. doi: 10.1093/nar/8.20.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel M., Model P. Characterization of the cloned fip gene and its product. J Bacteriol. 1984 Feb;157(2):526–532. doi: 10.1128/jb.157.2.526-532.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenbergh P. H., Sussenbach J. S., Roberts R. J., Jansz H. S. The 3'-terminal nucleotide sequences of adenovirus types 2 and 5 DNA. J Virol. 1975 Feb;15(2):268–272. doi: 10.1128/jvi.15.2.268-272.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J. I., Ohmori H., Bird R. E. Origin of replication of colicin E1 plasmid DNA. Proc Natl Acad Sci U S A. 1977 May;74(5):1865–1869. doi: 10.1073/pnas.74.5.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurushita N., Hirano M., Shigesada K., Imai M. Isolation and characterization of rho mutants of Escherichia coli with increased transcription termination activities. Mol Gen Genet. 1984;196(3):458–464. doi: 10.1007/BF00436193. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace B. J., Kushner S. R. Genetic and physical analysis of the thioredoxin (trxA) gene of Escherichia coli K-12. Gene. 1984 Dec;32(3):399–408. doi: 10.1016/0378-1119(84)90015-5. [DOI] [PubMed] [Google Scholar]
- Ward D. F., Gottesman M. E. The nus mutations affect transcription termination in Escherichia coli. Nature. 1981 Jul 16;292(5820):212–215. doi: 10.1038/292212a0. [DOI] [PubMed] [Google Scholar]
- Wu A. M., Christie G. E., Platt T. Tandem termination sites in the tryptophan operon of Escherichia coli. Proc Natl Acad Sci U S A. 1981 May;78(5):2913–2917. doi: 10.1073/pnas.78.5.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Crombrugghe B., Busby S., Buc H. Cyclic AMP receptor protein: role in transcription activation. Science. 1984 May 25;224(4651):831–838. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]