Abstract
Dolichol phosphate mannose (Dol-P-Man), formed upon transfer of Man from GDPMan to Dol-P, is a mannosyl donor in pathways leading to N-glycosylation, glycosyl phosphatidylinositol membrane anchoring, and O-mannosylation of protein. Dol-P-Man synthase is an essential protein in Saccharomyces cerevisiae. We have cloned cDNAs encoding human and Schizosaccharomyces pombe proteins that resemble S. cerevisiae Dol-P-Man synthase. Disruption of the gene for the S. pombe Dol-P-Man synthase homolog, dpm1+, is lethal. The known Dol-P-Man synthase sequences can be divided into two classes. One contains the S. cerevisiae, Ustilago maydis, and Trypanosoma brucei enzymes, which have a COOH-terminal hydrophobic domain, and the other contains the human, S. pombe, and Caenorhabditis synthases, which lack a hydrophobic COOH-terminal domain. The two classes of synthase are functionally equivalent, because S. cerevisiae DPM1 and its human counterpart both complement the lethal null mutation in S. pombe dpm1+. The findings that Dol-P-Man synthase is essential in yeast and that the Ustilago and Trypanosoma synthases are in a different class from the human enzyme raise the possibility that Dol-P-Man synthase could be exploited as a target for inhibitors of pathogenic eukaryotic microbes.
Dolichol phosphate mannose (Dol-P-Man), which is formed by transfer of mannose from GDPMan to the polyisoprenoid Dol-P, is a mannosyl donor in the assembly of the precursor oligosaccharide in N-glycosylation, in the synthesis of glycosylphosphatidylinositol anchors, and in O-mannosylation of fungal protein (1–3). Dol-P-Man seems to be synthesized at the cytoplasmic face of the membrane of the endoplasmic reticulum (ER) (4, 5), yet the Dol-P-Man-requiring steps in glycosylation are believed to take place in the ER lumen (5). Thus, Dol-P-Man supply may require not only a synthase but also proteins that translocate it into the ER lumen. Indeed, at least three genes may be involved in Dol-P-Man supply in mammals. Candidates for the synthase gene itself are the loci defective in the Thy-1 class E murine lymphoma mutant, the Chinese hamster ovary cell B4–2-1/Lec15 mutant, and a murine T cell hybridoma mutant, which are all defective in Dol-P-Man synthesis (6–8). The Lec35 gene may encode a protein that supplies Dol-P-Man to the ER lumen. Chinese hamster ovary cell Lec35 mutants make normal amounts of Dol-P-Man in vivo and in vitro but are unable to utilize it efficiently in vivo for glycosylation reactions, consistent with a defect in Dol-P-Man translocation (9). A third protein, SL15, may also be involved in Dol-P-Man supply, for the SL15 gene suppresses the mutations in both Dol-P-Man synthase-deficient Lec15 and Lec35 cells (10).
The Saccharomyces cerevisiae gene for Dol-P-Man synthase, DPM1, has been cloned and shown indeed to be the structural gene for the enzyme because it confers Dol-P-Man synthase activity on Escherichia coli when expressed in the bacteria (11). Dol-P-Man synthase genes from Trypanosoma brucei (12) and from Ustilago maydis (13) have been cloned, respectively, by complementation of a temperature-sensitive S. cerevisiae dpm1 mutant and on account of their cross-hybridization with S. cerevisiae DPM1. Although the isolation of a mammalian Dol-P-Man synthase gene would help us understand how Dol-P-Man is supplied in mammalian cells, such a gene has yet to be cloned. Mammalian Dol-P-Man synthase, though, would be expected to resemble the S. cerevisiae enzyme, given that the yeast DPM1 gene complements Dol-P-Man synthase-deficient mammalian cell lines (8, 14). However, it has been noted that the S. cerevisiae and mammalian Dol-P-Man synthases differ from one another in in vitro properties such as their sensitivity to nonionic detergents and their ability to interact with phospholipid vesicles (15, 16). A comparison of the amino acid sequences of mammalian and S. cerevisiae Dol-P-Man synthases may reveal differences between these enzymes. To explore this possibility and to identify conserved domains in Dol-P-Man synthases, we isolated Dol-P-Man synthase genes from diverse organisms.
We identified DNA sequences in the databases that encode human, nematode, and Schizosaccharomyces pombe proteins resembling S. cerevisiae Dol-P-Man synthase, and we report here the cloning of the human and S. pombe Dol-P-Man synthase genes and the lethal disruption of the S. pombe dpm1+ gene. The available Dol-P-Man synthase sequences fall into two classes, one containing the S. cerevisiae, T. brucei, and U. maydis proteins and the other the human, S. pombe, and nematode enzymes. The two classes of Dol-P-Man synthase are functionally equivalent, for the S. cerevisiae and human Dol-P-Man synthase genes both complement a lethal null mutation in S. pombe dpm1+.
MATERIALS AND METHODS
Strains and Culture Media.
The wild-type S. cerevisiae strain used was PP-1B (his4 chs1::URA3 ura3–52 MATa). Strain JZY251 (dpm1::LEU2/DPM1 ura3–52/ura3–52 leu2–3, 112/leu2–3, 112 MATa/MATα) (17) was used to test for rescue of the S. cerevisiae dpm1 disruption. S. pombe strains were derived from the heterothallic wild-type strains CHP 428 (h+ ade6-M210 leu1–32 his7–366 ura4-d18) and CHP 429 (h− ade6-M216 leu1–32 his7–366 ura4-d18). Growth media for S. cerevisiae were as described in ref. 11. S. pombe cells were grown in yeast extract liquid medium or in Edinburgh minimal medium 2 (EMM2) containing 100–250 μg/ml of supplements as required (18) and grown at 30°C.
Database Searching and Sequence Analyses.
The amino acid sequence of S. cerevisiae Dol-P-Man synthase (11) was used as a probe to search the dbEST (expressed sequence tag) and S. pombe genomic DNA databases using the tblastn algorithm (19). Sequences were analyzed using the dna strider 1.1 (20) and clustal v (21) programs. Hydropathy analyses were performed according to Kyte and Doolittle (22) and Rost et al. (23) and by using the toppred ii program (24).
PCR Amplification.
PCR amplification using specific and degenerate oligonucleotide primers was carried out using a succession of descending annealing temperatures to minimize nonspecific annealing of primers to template. Reaction mixtures for amplification by Vent DNA polymerase (New England Biolabs) contained 1 μM primers, 200 μM dNTPs, 10 μl of 10× Vent polymerase buffer, 4 mM MgSO4, and H2O in 98 μl. The DNA templates used contained 0.1–0.2 μg of plasmid DNA isolated using the Qiagen (Chatsworth, CA) “mini-prep” procedure or 1.3 μl of genomic DNA. Amplification by Taq DNA polymerase (BRL) was carried out following the same procedures but using 10× Taq DNA polymerase buffer and 6 mM MgCl2 instead of MgSO4. Samples were given a “hot start” at 94°C for 5 min and then maintained at 80°C for 1 min, after which polymerase was added. Amplification was then carried out using two linked cycles of 94°C for 1 min, 65°C for 1 min, and 72°C for 1 min, followed by two cycles under the same conditions except that the annealing temperature of 65°C was lowered to 62°C. These cycles were in turn followed by repetitions of two cycles in which the annealing temperature was successively lowered by 2°C until a temperature of 50°C was reached, at which temperature amplification was continued for 15 more cycles. A final cycle was performed at 94°C for 1 min, 50°C for 1 min, and 72°C for 3 min.
Isolation of Human and Rat Dol-P-Man Synthase cDNAs.
The oligonucleotides 5′-CGCGGATCCAAAAAGTTGGGACTAGGAACTGCA-3′ and 5′-CGCGGATCCTTTGTAGCGAGTTCCAGAGAC-3′ were used as forward and reverse primers respectively to amplify a 160-nucleotide fragment of Dol-P-Man synthase DNA from a human cDNA library in the vector pFL61 (26). The library was introduced into E. coli cells by electroporation, and a bacterial colony harboring a plasmid with the human Dol-P-Man synthase cDNA was identified by PCR screening of successive dilutions of suspensions of the bacterial transformants (27). A rat Dol-P-Man synthase cDNA was isolated by a combination of degenerate PCR, PCR screening, and colony hybridization. Forward and reverse degenerate primers were 5′-CGCGGATCCATGGAYGCIGAYYTIWSICA-3′ and 5′-CCGGAATTCICCIARYTTISWYTCICC-3′, respectively. PCR was performed using Taq DNA polymerase and a rat cDNA library (28) as template. A 370-nucleotide fragment was amplified and sequenced. The exact match reverse primer 5′-AAATCGGAACCTCACCAAC-3′ was used with the original degenerate forward primer to screen successive dilutions of suspensions of bacteria transformed with the rat cDNA library. A positive clone was localized to a pool of 10,000 transformants and then identified by colony hybridization (25) using the 370-nucleotide fragment as a probe. A full-length cDNA encoding the Caenorhabditis briggsiae Dol-P-Man synthase was provided by M. Marra (Genome Sequencing Center, Washington University, St. Louis, MO). Standard procedures were used for propagation and selection of plasmids, for growth of bacteria, for subcloning of DNA fragments, and for DNA sequencing (25).
Isolation of the S. pombe dpm1+ Gene.
The S. pombe genomic DNA sequence encoding a candidate Dol-P-Man synthase was present on cosmid c31G5, whose sequence was determined by J. McLean and D. Harris at the Sanger Centre, Hinxton, U.K. (personal communication). Oligonucleotides 5′-AAATATAGCGTTTTATTACCAACC-3′ and 5′-CATTTGAAATACATAACCTTTGCT-3′, matches to the genomic sequence, were used as forward and reverse primers respectively to amplify a 561-bp fragment of dpm1+ cDNA using an S. pombe cDNA library in the vector pFL61 (26) as template. The library was transformed into E. coli; the bacteria were plated on Luria–Bertani-ampicillin medium; and 17,000 colonies were screened by colony hybridization (25) using the [α-32P]dCTP-labeled 561-bp fragment as probe.
Vector Constructs.
Forward and reverse PCR primers with NdeI and BamHI sites at their respective 5′ ends were used to amplify cDNAs for subcloning into the S. pombe expression vector pREP2 (29). Since the human Dol-P-Man synthase cDNA lacked an initiation codon, one was added by engineering an ATG codon in the NdeI site of the forward primer immediately 5′ to the native arginine codon. Human and S. cerevisiae Dol-P-Man synthases were amplified by PCR using as template 0.1–0.2 μg of plasmid with the appropriate cDNA that had been isolated using the Qiagen “mini-prep” procedure. S. pombe dpm1+ was amplified using 1.3 μg of genomic DNA as template. PCR was carried out using Vent polymerase, and amplified DNA was then digested with NdeI and BamHI and ligated into pREP2. The PCR-amplified human Dol-P-Man synthase gene with an added 5′ ATG codon was also subcloned into a galactose-inducible 2 μm S. cerevisiae expression vector.
Disruption of S. pombe dpm1+.
Primers SpDDF3 and SpDDR3, 5′-CCGGAATTCCGTCAAGTTCGTCCTACTGCTCCT-3′ and 5′-CGGGGTACCGGTTGGTAATAAAACGCTATATTTTGA-3′, respectively, and Taq polymerase were used to amplify a DNA fragment containing the first 30 nucleotides of the S. pombe dpm1+ coding region and 1 kb of upstream DNA. This fragment was cloned into the EcoRI and KpnI sites of pNEB193 (New England Biolabs) using restriction sites engineered into the primers, to give plasmid pNDD1. Primers SpDDF4 and SpDDR4, 5′-CGCGGATCCTTGCCTTTGTCGACAGATTGTATGGTG-3′ and 5′AAGGTTACGTAACCACTGCAGTTGTTGGAGGATCAATTTGTATCTCAC-3′, respectively, were used to amplify a DNA fragment consisting of 95 bases at the 3′ end of S. pombe dpm1+ and 716 bases of adjacent 3′ DNA. This fragment was cloned into the BamHI and PstI sites of pNND1 using sites engineered into SpDDF4 and SpDDR4 to give pNDD2. A KpnI-BamHI DNA fragment containing the S. pombe his7+ gene (30) was then subcloned into the KpnI and BamHI sites of pNND2 to give plasmid pΔdpm1+. A linear DNA fragment containing the disrupted S. pombe dpm1+ gene was amplified by PCR using primers SpDDF3 and SpDDR4 and plasmid pΔdpm1+ as template and was used to transform the his7−/his7− S. pombe diploid strain 428 × 429. Histidine prototrophs were selected on minimal medium, and a colony PCR screen (31) was carried out on 27 transformants to identify those that had integrated the disrupting fragment at the dpm1+ locus. For this, primers were designed to anneal to the his7+ gene close to the BamHI site of the disrupting fragment and to genomic DNA further downstream of that used to make that fragment. These oligonucleotides would prime amplification of a 1-kb fragment of DNA from only those diploid colonies that contain the disrupting fragment at the dpm1+ locus. A DNA fragment of the expected size was amplified from 2 of the 27 histidine prototrophic diploids, and one, dpm1+::his7+/dpm1+.S27, was studied further.
Random Spore Analysis.
Strain dpm1+::his7+/dpm1+.S27 was allowed to sporulate on EMM2 medium containing appropriate supplements. Asci was transferred to EMM2 agar plates that either lacked histidine or that were supplemented with 100 mg of histidine per liter, and ascospores were allowed to germinate. Absence of any haploid colonies on medium lacking histidine would indicate that the dpm1+::his7+ disruption is lethal. Haploid colonies were distinguished from diploid colonies by the ade6-M210/ade6-M216 intrallelic complementation system (18).
Complementation of the dpm1+::HIS7+ Disruption.
The human, S. cerevisiae and S. pombe Dol-P-Man synthase genes were amplified by PCR and cloned into the S. pombe expression vector pREP2 (29) to give plasmids pHsDPM1, pScDPM1, and pSpdpm1+. These plasmids, as well as pREP2 alone, were transformed into diploid dpm1+::his7+/dpm1+.S27. Uracil prototrophs were induced to sporulate and asci were then transferred to 5 ml of water containing 0.5% (vol/vol) Glusulase (DuPont), in which they were incubated overnight at 30°C to allow Glusulase to digest ascal walls and destroy vegetative cells. About 4,000 spores from each diploid were spread on medium selective for histidine prototrophs.
Dol-P-Man Synthase Assays.
dpm1+::his7+ haploids harboring pSpdpm1+, pScDPM1, or pHsDPM1 were grown in medium selective for histidine and uracil prototrophy and harvested at mid-log phase. Membrane fractions were prepared from cells of each culture as described in ref. 32, except that protease inhibitors were omitted. Transfer of [14C]Man from GDP[14C]Man to endogenous Dol-P in S. pombe membranes was measured according to procedures detailed in ref. 11.
RESULTS
Isolation of Dol-P-Man Synthase Homologs.
Dol-P-Man synthase has a key role in the assembly of the carbohydrate and glycolipid substituents of eukaryotic cell surface proteins, yet little is known of the structure and function of this enzyme. To identify conserved domains of the protein, we compared Dol-P-Man synthases from diverse eukaryotes. Human and C. briggsiae cDNAs encoding protein fragments resembling S. cerevisiae Dol-P-Man synthase were found in the dbEST database and a candidate fission yeast Dol-P-Man synthase gene was found in the S. pombe genomic DNA database. Human and rat cDNAs encoding putative Dol-P-Man synthases were then isolated from cDNA libraries and sequenced. Neither the human cDNA, nor its rat counterpart, which encodes a protein 93% identical to human Dol-P-Man synthase, had a methionine codon at their 5′ end, and both cDNAs are presumably derived from truncated mRNAs. However, as described below, the human cDNA with an ATG codon added at its 5′ end encodes a functional protein; thus, should the human enzyme be longer at its NH2 terminus, that part of the protein is nonetheless dispensible for function in vivo.
A cDNA for S. pombe Dol-P-Man synthase was identified by using a PCR-amplified fragment of the gene to screen an S. pombe cDNA library by colony hybridization. The genomic S. pombe dpm1+ DNA contains a 119-nucleotide intron toward its 3′ end. The intron is flanked by 5′ and 3′ splice sites and contains an internal branch site; all three are exact matches to published S. pombe splice site consensus sequences (33).
The predicted human, S. pombe, and C. briggsiae Dol-P-Man synthases contain, respectively, 253, 236, and 242 amino acids (Fig. 1). Conserved in the three new synthases are sequences that are close matches to Val-Asp-Asp-Asn-Ser and Met-Asp-Ala-Asp-Leu in the S. cerevisiae enzyme. The italicized aspartic acid residues in these sequences (Fig. 1) are conserved in a range of β-glycosyltransferases that transfer a single sugar residue, and these aspartates have been suggested to be involved in catalysis (34). The new Dol-P-Man synthases have a serine residue in the position corresponding to Ser-141 in the S. cerevisiae enzyme (Fig. 1). This serine, and the conserved residues NH2-terminal to it, meet certain criteria for a consensus site for phosphorylation by cAMP-dependent kinase, and indeed, Dol-P-Man synthase may be phosphorylated by this enzyme (35). None of the proteins contains a sequence of hydrophobic amino acids resembling that found in S. cerevisiae Dol-P-Man synthase and suggested to be involved in dolichol recognition (36). The new candidate Dol-P-Man synthases are unlikely to be Dol-P-Glc synthases, for we have also cloned a human counterpart of S. cerevisiae Dol-P-Glc synthase and shown that it is enzymatically active when expressed in E. coli (C.T and P.O., unpublished data).
Figure 1.
Alignment of the amino acid sequences of Dol-P-Man synthases. Sequences were aligned using the clustal v program (21). Amino acids identical in at least four of the six sequences are highlighted in black and similar residues are highlighted in gray. The aspartic acid residues marked ∗ are conserved in β-glycosyltransferases that transfer a single sugar residue and which may be involved in catalysis (34). The serine marked + and the amino acids NH2-terminal to it meet certain criteria for a consensus site for phosphorylation by cAMP-dependent kinase. Abbreviations and references to published sequences are as follows: Sc, S. cerevisiae (11); Um, U. maydis (12); Tb, T. brucei (13); Hs, human; Cb, C. briggsiae; Sp, S. pombe.
Identification of Two classes of Dol-P-Man Synthase.
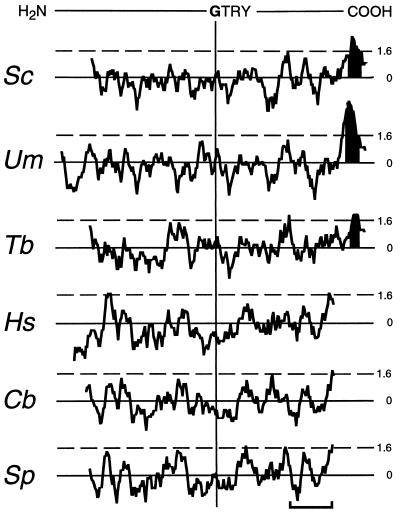
Alignment of the S. cerevisiae, U. maydis and T. brucei Dol-P-Man synthases with their human, fission yeast, and nematode homologs using the clustal v algorithm reveals that the six proteins show 39% overall identity plus similarity (relative to the shortest sequence) (Fig. 1). However, the proteins can be divided into two distinct groups. One group contains the S. cerevisiae, U. maydis, and T. brucei enzymes, whose amino acid sequences show 61% overall identity plus similarity, and the other contains the human, S. pombe, and C. briggsiae proteins, 74% of whose amino acid residues are either identical or similar to one another. The fact that S. pombe Dol-P-Man synthase shows greater similarity to the human enzyme is consistent with observations that S. pombe resembles mammalian cells more closely in certain aspects of its cellular biochemistry than it resembles S. cerevisiae (37). Proteins of the S. cerevisiae class are some 30 amino acids longer at their COOH terminus than human class proteins. The differences between the two classes of Dol-P-Man synthase are also apparent when their hydropathy plots (22) are aligned using the sequence Gly-Thr-Arg-Tyr as a reference (Fig. 2); this shows that proteins within each class have similar hydropathy profiles but that the two classes differ from one another. Strikingly, members of the human class of protein lack the hydrophobic COOH terminus that is characteristic of the S. cerevisiae class of Dol-P-Man synthase. Moreover, members of the human class of synthase do not contain any stretches of amino acids capable of forming a transmembrane domain according to the program of Rost et al. (23). This program, however, identifies a single potential transmembrane helix at the COOH termini of the S. cerevisiae, T. brucei, and U. maydis proteins (Fig. 2) and, furthermore, predicts that the bulk of each protein of the S. cerevisiae class has a cytoplasmic orientation. No “certain” membrane-spanning segments were identified in the human class Dol-P-Man synthases by the toppred ii analysis program (24). The latter analysis, however, predicts the existence of a “certain” membrane-spanning segment at the COOH terminus of each of the S. cerevisiae class enzymes.
Figure 2.
Hydropathy plots of Dol-P-Man synthases. Plots were generated according to Kyte and Doolittle (22) with a window of 11, using the dna strider 1.1 program (20). Plots are aligned to the Gly-Thr-Arg-Tyr (GTRY) sequence conserved in all Dol-P-Man synthases. Stretches of amino acids with hydropathies above 1.6 (---) may form transmembrane domains. Transmembrane sequences predicted by the program of Rost et al. (23) are shaded in black. (Bar = 50 amino acids.)
S. cerevisiae and Human Class Dol-P-Man Synthases are Functionally Equivalent.
Given the difference between the human and S. cerevisiae class Dol-P-Man synthases, it was important to demonstrate that the human enzyme is functionally equivalent to the product of the S. cerevisiae DPM1 gene, which was shown unequivocally to be the structural gene for Dol-P-Man synthase by heterologous expression in E. coli (11). Unlike S. cerevisiae DPM1, however, the human Dol-P-Man synthase gene did not confer Dol-P-Man synthase activity on E. coli, and neither the human nor S. pombe gene restored viability to haploid S. cerevisiae dpm1::LEU2 disruptants.
Since human Dol-P-Man synthase is 65% identical to its S. pombe counterpart, and since S. cerevisiae DPM1 is functionally expressed in Dol-P-Man synthase-deficient mammalian cell lines (8, 14), we tested whether the dpm1+ gene is essential for viability in S. pombe and, if so, whether a lethal null mutation can be rescued by both the S. cerevisiae and human Dol-P-Man synthase genes. S. pombe dpm1+ was disrupted by replacing 50% of its coding region with the marker his7+. Diploid dpm1+::his7+/dpm1+.S27, which was confirmed by Southern blot analysis to have integrated the dpm1+::his7+ fragment at its chromosomal dpm1+ locus, was allowed to sporulate, and random spore analysis was used to determine the null phenotype of the dpm1+::his7+ disruption. No histidine-prototrophic haploid colonies grew, and we conclude that dpm1+ is essential for viability in S. pombe.
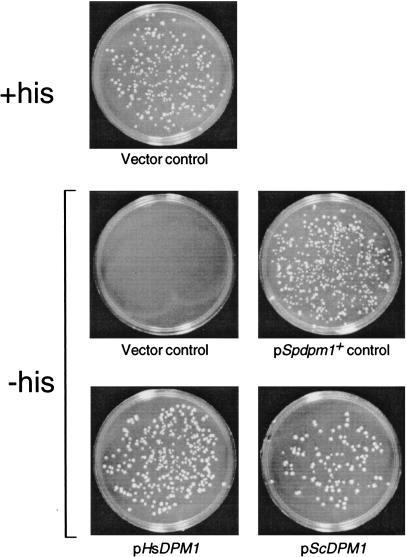
DNAs encoding the S. cerevisiae, human, and S. pombe Dol-P-Man synthases were subcloned into the ura4+-marked S. pombe expression vector pREP2 to test whether they complement the lethal dpm1+::his7+ null phenotype. Diploid dpm1+::his7+/dpm1+.S27 was transformed to uracil prototrophy with pScDPM1, pHsDPM1, or pSpdpm1+, and with pREP2 alone as a control. The transformed diploids were allowed to sporulate, and the meiotic progeny were submitted to random spore analysis to test whether haploid cells could grow on medium selective for both histidine and uracil prototrophy. Fig. 3 shows that introduction of plasmids containing the S. cerevisiae and human Dol-P-Man synthase genes, as well as S. pombe dpm1+, restored growth to dpm1+::his7+ haploids, whereas pREP2 alone did not.
Figure 3.
S. cerevisiae and human Dol-P-Man synthase genes allow growth of haploid S. pombe dpm1+::his7+ cells. Heterozygous dpm1+::his7+/dpm1+ diploids harboring pScDPM1, pHsDPM1, pSpdpm1+, or the vector pREP2 were allowed to sporulate, and the meiotic progeny were tested for growth on medium selective for histidine and uracil prototrophy. Segregants from the dpm1+::his7+/dpm1+ diploid harboring pREP2 were also plated on medium containing histidine to select for wild-type haploids containing pREP2.
Membranes prepared from the uracil plus histidine-prototrophic haploids all had in vitro Dol-P-Man synthase activity (not shown). These results show that the protein products of the human and S. pombe Dol-P-Man synthase genes are functionally equivalent to that of S. cerevisiae DPM1; the human class proteins are therefore indeed Dol-P-Man synthases. Three mammalian cell lines are defective in Dol-P-Man synthase: the Thy-1 class E murine lymphoma mutant (6), the Chinese hamster ovary cell mutant B4–2-1 (7), and a murine T cell hybridoma mutant (8). If the mutation in any of these cell lines is in the gene for Dol-P-Man synthase, then our human or rat cDNA would be expected to complement it.
DISCUSSION
Our finding that Dol-P-Man synthases fall into two classes raises issues as to how Dol-P-Man is synthesized in different eukaryotes, and also has practical applications in that differences between the Dol-P-Man synthases of humans and microbes could be exploited in the development of antimicrobial agents.
The amino acid sequence differences between the S. cerevisiae and human Dol-P-Man synthases, and the presence and absence, respectively, of a COOH-terminal transmembrane domain in these enzymes, are correlated with observations that the S. cerevisiae and mammalian Dol-P-Man synthases differ from one another. Thus, the two types of synthase differ in their sensitivity to nonionic detergents and in their interactions with phospholipids. Partially purified rat Dol-P-Man synthase associates with phospholipid vesicles when the latter contain Dol-P, and formation of this complex is necessary for enzymatic activity. In contrast, the bacterially expressed S. cerevisiae enzyme can associate with phospholipid vesicles in the absence of Dol-P, but this complex does not support enzyme activity (15, 16). Yeast and mammalian Dol-P-Man synthases may therefore differ in how they interact with membranes in vivo. S. cerevisiae and human class Dol-P-Man synthases also differ in their ability to be expressed in E. coli and to complement mutations in the other class of enzyme. Thus, the S. cerevisiae synthase is expressed as an active enzyme in E. coli, but the human enzyme is not, and whereas the S. cerevisiae DPM1 gene complements Dol-P-Man synthesis-defective mammalian cell lines, the human gene does not rescue the lethal S. cerevisiae dpm1 disruption.
The latter observations suggest that the S. cerevisiae synthase can function autonomously but that the human enzyme cannot. One explanation is that the human class enzyme requires one or more additional proteins for activity. A function for an auxiliary protein might be to promote membrane attachment of the synthase, which lacks an obvious transmembrane domain. However, since native mammalian and S. pombe Dol-P-Man synthases both sediment with membranes and are therefore normally membrane-associated, the auxiliary protein may instead be required for catalytic activity. The SL15 gene’s product is a candidate for an auxiliary protein: SL15 suppresses the mutation in mammalian Dol-P-Man synthase-deficient Lec15 cells (10). The SL15 protein, which shows no resemblance to Dol-P-Man synthases, has two transmembrane domains and a COOH-terminal ER retention signal, and could therefore interact with Dol-P-Man synthase at the ER membrane. Although SL-15 has no S. cerevisiae homolog, it may well have its counterpart in fission yeast, and mutants in S. pombe SL-15 might show defects in Dol-P-Man synthesis and in the membrane localization of Dol-P-Man synthase. As fission yeast Dol-P-Man synthase is 65% identical to its human counterpart and can be replaced by the human enzyme, an S. pombe strain expressing human Dol-P-Man synthase will be a useful model system in which to explore the Dol-P-Man supply machinery of mammalian cells. Moreover, the S. pombe expression system will permit a detailed comparison of the human and S. cerevisiae synthases, for example, with respect to how these proteins might differ in their association with membranes.
The similarities and differences between the two classes of Dol-P-Man synthase also have implications for how Dol-P is recognized. The results of protease-protection experiments (4) and of analyses using predictive algorithms (23) suggest that S. cerevisiae Dol-P-Man synthase faces the cytoplasm and is anchored in the ER membrane by its COOH-terminal hydrophobic domain. The human class of Dol-P-Man synthase presumably has the same membrane orientation as the S. cerevisiae enzyme, a notion supported by the fact that the S. cerevisiae enzyme is functional when expressed in mammalian cells (8, 14). Human class Dol-P-Man synthases lack any COOH-terminal hydrophobic sequence resembling that proposed to be involved in dolichol recognition (36). However, human class Dol-P-Man synthases, as well a truncated form of the S. cerevisiae enzyme lacking all but three of its COOH-terminal hydrophobic residues, function in vivo, indicating that a hydrophobic COOH-terminal transmembrane domain is not critical for recognition of Dol-P (17), an interaction that might be predicted to take place in the membrane.
Dol-P-Man synthase is essential in S. cerevisiae (11), and temperature-sensitive dpm1 mutants are blocked in steps in N-glycosylation, in O-mannosylation, and in glycosylphosphatidylinositol anchoring (38). Abolition of Dol-P-Man synthesis will therefore in turn prevent formation of key surface glycoproteins of eukaryotic microbes. It is striking that the Dol-P-Man synthases of two eukaryotic microbial pathogens, T. brucei and U. maydis, are in a different class from the human synthase. The possibility has been raised that microbial Dol-P-Man synthases may differ from the mammalian enzyme and be inhibited selectively by antiparasitic drugs (12, 16). Our demonstration that human Dol-P-Man synthase is indeed different from its known fungal and protozoal counterparts is powerful support for this notion. The fact that both S. cerevisiae and human class Dol-P-Man synthases function in S. pombe could be exploited in a differential screen for agents that inhibit the growth of S. pombe cells relying on a pathogen’s Dol-P-Man synthase, but not growth of S. pombe cells harboring the human enzyme.
Acknowledgments
We thank I. Mac Allister for helpful discussions; Drs. B. N. Marbois and C. F. Clarke for their rat cDNA library; Drs. M. Minet and F. Lacroute for their human and S. pombe cDNA libraries; Drs. B. Barrell, J. McLean, and D. Harris for S. pombe sequence information; and Dr. M. Marra for the C. briggsiae EST clone. We are also grateful to Dr. T. Kinoshita for sharing sequence information. The Sanger Centre is supported by the Wellcome Trust. This work was supported by National Institutes of Health Grant GM46220.
ABBREVIATIONS
- Dol-P-Man
dolichol phosphate mannose
- ER
endoplasmic reticulum
- EMM2
Edinburgh minimal medium 2
Footnotes
References
- 1.Kornfeld R, Kornfeld S. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 2.Tanner W, Lehle L. Biochim Biophys Acta. 1987;906:81–99. doi: 10.1016/0304-4157(87)90006-2. [DOI] [PubMed] [Google Scholar]
- 3.Herscovics A, Orlean P. FASEB J. 1993;7:540–550. doi: 10.1096/fasebj.7.6.8472892. [DOI] [PubMed] [Google Scholar]
- 4.Clarke B L, Naylor C, Lennarz W J. Chem Phys Lipids. 1989;51:239–247. doi: 10.1016/0009-3084(89)90011-x. [DOI] [PubMed] [Google Scholar]
- 5.Abeijon C, Hirschberg C B. Trends Biochem Sci. 1992;17:32–36. doi: 10.1016/0968-0004(92)90424-8. [DOI] [PubMed] [Google Scholar]
- 6.Chapman A, Fujimoto K, Kornfeld S. J Biol Chem. 1980;255:4441–4446. [PubMed] [Google Scholar]
- 7.Stoll J, Robbins A R, Krag S S. Proc Natl Acad Sci USA. 1982;79:2296–2300. doi: 10.1073/pnas.79.7.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeGasperi R, Thomas L J, Sugiyama E, Chang H M, Beck P, Orlean P, Albright C, Waneck G, Sambrook J F, Warren C D, Yeh E T H. Science. 1990;250:988–991. doi: 10.1126/science.1978413. [DOI] [PubMed] [Google Scholar]
- 9.Zeng Y, Lehrman M A. J Biol Chem. 1990;265:2296–2305. [PubMed] [Google Scholar]
- 10.Ware F E, Lehrman M A. J Biol Chem. 1996;271:13935–13938. doi: 10.1074/jbc.271.24.13935. [DOI] [PubMed] [Google Scholar]
- 11.Orlean P, Albright C, Robbins P W. J Biol Chem. 1988;263:17499–17507. [PubMed] [Google Scholar]
- 12.Mazhari-Tabrizi R, Eckert V, Blank M, Müller R, Mumberg D, Funk M, Schwarz R T. Biochem J. 1996;316:853–858. doi: 10.1042/bj3160853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zimmerman J W, Specht C A, Cazares B X, Robbins P W. Yeast. 1996;12:765–771. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C765::AID-YEA974%3E3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 14.Beck P J, Orlean P, Albright C, Robbins P W, Gething M-J, Sambrook J F. Mol Cell Biol. 1990;10:4612–4622. doi: 10.1128/mcb.10.9.4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen J W, Schutzbach J S. Biochemistry. 1988;27:6315–6320. doi: 10.1021/bi00417a017. [DOI] [PubMed] [Google Scholar]
- 16.Schutzbach J S, Zimmerman J W, Forsee T W. J Biol Chem. 1993;268:24190–24196. [PubMed] [Google Scholar]
- 17.Zimmerman J W, Robbins P W. J Biol Chem. 1993;268:16747–16753. [Google Scholar]
- 18.Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. Experiments with Fission Yeast. A Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. [Google Scholar]
- 19.Altschul S F, Gish W, Miller W, Myers E, Lipman D. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 20.Marck C. Nucleic Acids Res. 1988;16:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higgins D G. In: Methods in Molecular Biology: Computer Analysis of Sequence Data, Part II. Griffin A M, Griffin H G, editors. Totowa, NJ: Humana; 1994. [DOI] [PubMed] [Google Scholar]
- 22.Kyte J, Doolittle R F. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 23.Rost B, Casadio R, Fariselli P, Sander C. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sipos L Von Heijne. Eur J Biochem. 1993;213:1333–1340. doi: 10.1111/j.1432-1033.1993.tb17885.x. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab.; 1989. [Google Scholar]
- 26.Minet M, Dufour M-E, Lacroute F. Plant J. 1992;2:417–422. doi: 10.1111/j.1365-313x.1992.00417.x. [DOI] [PubMed] [Google Scholar]
- 27.Takumi T, Lodish H F. BioTechniques. 1994;17:443–444. [PubMed] [Google Scholar]
- 28.Marbois B N, Hsu A, Pillai R, Colicelli J, Clarke C F. Gene. 1994;138:213–217. doi: 10.1016/0378-1119(94)90810-9. [DOI] [PubMed] [Google Scholar]
- 29.Maundrell K. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- 30.Apolinario E, Nocero M, Jin M, Hoffman C S. Curr Genet. 1993;24:491–495. doi: 10.1007/BF00351711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jesnowski R, Naehring J, Wolf K. Curr Genet. 1995;27:318–319. doi: 10.1007/BF00352100. [DOI] [PubMed] [Google Scholar]
- 32.Colussi P A, Orlean P. Yeast. 1997;13:139–150. doi: 10.1002/(SICI)1097-0061(199702)13:2<139::AID-YEA69>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 33.Nasim A, Young P, Johnson B F. Molecular Biology of the Fission Yeast. San Diego: Academic; 1989. [Google Scholar]
- 34.Saxena I M, Brown R M, Fevre M, Geremia R A, Henrissat B. J Bacteriol. 1995;177:1419–1424. doi: 10.1128/jb.177.6.1419-1424.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banerjee D K, Kousvelari E E, Baum B J. Proc Natl Acad Sci USA. 1987;84:6389–6393. doi: 10.1073/pnas.84.18.6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albright C F, Orlean P, Robbins P W. Proc Natl Acad Sci USA. 1989;86:7366–7369. doi: 10.1073/pnas.86.19.7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Y, Lieberman H B. DNA Cell Biol. 1995;14:359–371. doi: 10.1089/dna.1995.14.359. [DOI] [PubMed] [Google Scholar]
- 38.Orlean P. Mol Cell Biol. 1990;10:5796–5805. doi: 10.1128/mcb.10.11.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]