Abstract
Coactivators, such as steroid receptor coactivator 1 (SRC-1A) and CREB (cAMP response element binding protein)-binding protein (CBP), are required for efficient steroid receptor transactivation. Using an in vitro transcription assay, we found that progesterone receptor (PR)-driven transcription is inhibited by a dominant negative PR ligand-binding domain-interacting region of SRC-1A, indicating that SRC-1A is required for actual transcriptional processes. In addition, these coactivators also possess intrinsic histone acetyltransferase (HAT) activity and bind to each other and another HAT, p300/CBP-associated factor. Here we show that the human PR also interacts with p300/CBP-associated factor in vitro. Recruitment of multiple HATs to target promoters suggests an important role for chromatin remodeling in transcriptional activation of genes by steroid receptors. In transient transfection assays, we found that addition of a histone deacetylase inhibitor, trichostatin A, strongly potentiated PR-driven transcription. In contrast, directing histone deacetylase-1 (HD1) to a promoter using the GAL4 DNA binding domain inhibited transcription. Furthermore, PR transactivation was repressed by recruiting HD1 into the PR-DNA complex by fusing HD1 to a PR ligand-binding domain-interacting portion of SRC-1. Collectively, these results suggest that targeted histone acetylation by recruited HAT cofactors and histone deacetylation are important factors affecting PR transactivation. Recruitment of coactivators and HATs by the liganded PR in vivo may result in (i) remodeling of transcriptionally repressed chromatin to facilitate assembly and (ii) enhanced stabilization of the preinitiation complex by the activation functions of coactivators and the liganded PR itself.
Steroid hormones exert profound effects on cell growth, development, differentiation, and homeostasis. Their effects are mediated through specific intracellular steroid receptors that act as hormone-dependent transcription factors. Upon ligand binding, these receptors are able to recognize specific hormone response elements located in or near promoter DNA regions of target genes, resulting in positive or negative effects on transcription (reviewed in refs. 1 and 2). Several models have been proposed to explain the mechanism(s) by which steroid receptors activate gene transcription in vivo. One model implicates chromatin remodeling in steroid receptor transactivation (reviewed in refs. 3 and 4). A second model involves direct binding of steroid receptors to proteins in the preinitiation complex (PIC) such as TFIIB (5–7). These direct protein–protein interactions would result in stabilization of PIC assembly and increased rates of transcription initiation. A third model proposes that steroid receptors work through complexing proteins or steroid receptor coactivators to enhance assembly of a stable PIC. This idea was supported initially by squelching experiments in which different liganded steroid receptors inhibited transactivation by each other, suggesting the sequestering of a common, limiting, and essential factor(s) (8, 9).
Recently, several coactivators have been cloned and characterized that associate with steroid receptors and enhance their ability to transactivate target genes (reviewed in refs. 10 and 11). Given that these coactivators have intrinsic activation functions, these factors most likely enhance assembly of basal transcription factors into a stable PIC, resulting in increased transcription initiation rates of RNA polymerase II (12, 13). Surprisingly, the coactivators CREB (cAMP response element-binding protein)-binding protein (CBP) and steroid receptor coactivator 1 (SRC-1A) have been found to possess intrinsic histone acetyltransferase (HAT) activities, in addition to several putative activation domains (refs. 14 and 15; T.E.S., G.J., C. D. Allis, J. Zhou, C. A. Mizzen, N. J. McKenna, S. A. Onate, S.Y.T., M.-J.T., and B.W.O., unpublished work). Moreover, CBP and SRC-1A interact with each other and another HAT, p300/CBP-associated factor (PCAF) (ref. 17; T.E.S. et al., unpublished work). Thus, steroid receptor transactivation of target genes in vivo also may involve chromatin remodeling through targeted histone acetylation by recruited coactivators.
Genomic DNA and episomal DNA, introduced by transient transfection in cell culture, have been shown to package into nucleosomal structures or arrays (18, 19). Nucleosomes consist of an octamer of histones, H2A, H2B, H3, and H4, around which 146 bp of double-helix DNA is wound. Nucleosomes are potent repressors of transcription both in vitro and in vivo (20–22). Assembly of DNA into nucleosomes generally inhibits transcription factor binding, represses basal promoter activity, and is required for the proper regulation of many inducible genes (23). Nucleosome disruption is an essential regulatory step in the transactivation of many inducible genes by the nuclear receptors (24, 25). Depending on the positioning of cognate DNA response elements, this structure can place a strong repressive force on gene transcription by precluding transcription factor access to their DNA elements (26, 27). Targeted histone acetylation is thought to neutralize the positive charge of the histone N termini and weaken or “loosen” the interaction between histones and the negatively charged DNA. Loosening of the nucleosomal structure should increase access of transcription factors to their cognate DNA response elements in target gene promoter regions. Nucleosome displacement by histone acetylation can be reversed by histone deacetylation (26, 28, 29). The first histone deacetylase (HD1) was recently cloned (30). Trichostatin A (TSA), a potent inhibitor of histone deacetylation, has been shown to affect gene transactivation resulting in alteration of cell cycle kinetics (31). Therefore, the activity of promoters in vivo may reflect a competition between histone acetylation and deacetylation. Thus, targeted histone acetylation by transcription factors and recruited cofactors probably is a major step in steroid transactivation of gene expression in transcriptionally repressed chromatin.
To investigate the importance of coactivation and chromatin remodeling in transactivation by the human progesterone receptor (PR), we first used an in vitro transcription system to substantiate the necessity of the activation functions of SRC-1A for PR transactivation independent of histone modification. Next, we demonstrated that the human PR directly interacts with PCAF, suggesting that many HATs are recruited into a DNA complex when the PR is bound by ligand. We then determined that PR transactivation is affected by both inhibition of histone deacetylases as well as recruitment of HD1 in the PR complex. Collectively, results presented here indicate that both chromatin remodeling and transcriptional activation are essential but perhaps separate processes required for PR transactivation.
MATERIALS AND METHODS
Materials.
R5020 (promegestone) was purchased from NEN. RU486 was from Roussel-UCLAF. TSA was from Wako Pure Chemical (Osaka).
Plasmid DNAs.
Mammalian expression vectors for human PRB (32) and Flag-PCAF (PCR3.1Flag-PCAF) (T.E.S. et al., unpublished work), baculovirus expression plasmids pVLGST and pVLGST-hPRA (40), the bacterial expression plasmids pRSETGST-SRC-1(1–399) and pRSETGST-SRC-1(384–842) (T.E.S. et al., unpublished work), the yeast expression plasmid pCBGST-SRC-1(1216–1441) (T.E.S. et al., unpublished work), progesterone response element (PRE)2-TATA-G-free and adenovirus major late promoter (AdML)-G-free reporters (39), and (UAS)4-tk-LUC (LUC, luciferase) reporter (33) have been described. The PR-responsive (PRE)2-E1b-LUC reporter was constructed by subcloning the HindIII–BglII fragment of (PRE)2-E1b-CAT (CAT, chloramphenicol acetyltransferase) into the pGL3 vector (Promega). The yeast pCBGST, pCBGSThPR1–164, pCBGSThPR165–535, and pCBGSThPR631–933 expression vectors were from S. A. Onate (Baylor College of Medicine).
The SRC-1A N terminus (amino acids 1–360) was constructed by PCR from a human heart library using the following primers: 5′-CATCATCATGAGTGGCCTTGGGG-3′, encompassing the ATG start site of mouse SRC-1A (34), and 5′-GGATTGACCGAGGGATTTACTCGG-3′ from the original human SRC-1 clone (35). The PCR product encoding amino acids 1–399 of human SRC-1A was inserted into self-ligated PCR3.1 T/A cloning vector (Invitrogen). PCR3.1hSRC-1A was constructed from PCR3.1hSRC-1A1–399 by inserting the 3.3-kb partial AvaI–XbaI SRC-1 fragment from pBKCMVSRC-1 (35) into the AvaI–XbaI-digested PCR3.1hSRC-1A1–399 vector. The HD1-F cDNA was obtained from S. L. Schreiber (Howard Hughes Medical Institute, Cambridge, MA) and subcloned into pABGAL1–147 (36) by SalI and Klenow filled-in BamHI digestion of pBJHD-1 (30) insertion in XbaI-filled-in and SalI-digested pABGAL1–147 vector. PCR3.1HD1 mammalian expression plasmid was constructed by transferring HD1 cDNA from BamHI-filled-in pABGAL1–147 HD1 to PCR3.1 vector digested with AflII–XbaI and filled in with Klenow enzyme, PCR3.1SRC-1(1180–1441) was constructed by inserting the BspHI-filled-in/NdeI plasmid fragment from PCR3.1 hSRC-1A into the HindIII-filled-in/NdeI PCR3.1hSRC-1A vector. PCR3.1HD1-SRC-1(1180–1441) was constructed by inserting the BspHI-filled-in/XbaI fragment from PCR3.1hSRC-1A into the SalI-filled-in/XbaI PCR3.1HD1 vector. Cytomegalovirus-GL914HD1-SV was constructed by inserting the BamHI-filled-in HD1 full-length cDNA from PCR3.1-HD1 into the SalI–BamHI-digested and filled-in cytomegalovirus-GL914VPc-SV (37).
Cell-Free in Vitro Transcription Assay.
Proteins encoded by pCBGST and pCBGST-SRC-1(1216–1441) plasmids were expressed in the BJ2168 yeast cell line and isolated as described (38). Proteins were purified by binding to glutathione-Sepharose beads, extensive washing with TB buffer (20 mM Hepes, pH 7.9/20% glycerol/100 mM KCl/0.2 mM EDTA/5 mM MgCl2/2 mM dithiothreitol), and eluted with 100 mM glutathione in TB. Glutathione was removed, and proteins were concentrated using centrifugal concentrators (Filtron, Karlstein, Germany). Protein concentration was determined by Bradford assay, and purity was assessed by SDS/PAGE. The in vitro transcription assay was performed as described with few modifications (39). Instead of the proteinase K digestion, the 30-μl reaction was terminated with 200 μl of stop mix (0.5% SDS/10 mM Tris, pH 8.0/10 mM EDTA/0.3 M NaAc, pH 5/6 M urea). Radioactive bands were quantitated using a Betagen (Waltham, MA) blot scanner.
In Vitro Protein-Binding Assay.
The glutathione S-transferase (GST) and GST-hPRA were expressed in Sf9 insect cells by baculovirus infection (40). The GST-hPR1–164, GST-hPR165–535, and GST-hPR631–933 were expressed in yeast cells (38). These fusion proteins were bound to glutathione-Sepharose beads and washed with NETN100 (20 mM Tris⋅HCl, pH 8.0/1 mM EDTA, pH 8.0/0.5% Nonidet P-40/100 mM NaCl). Beads were then incubated for 1 hr at room temperature with 35S-labeled Flag-PCAF protein in vitro transcribed and translated from PCR3.1 Flag-PCAF using the Promega TnT kit. Beads were extensively washed with NETN100, and the bound material was analyzed by SDS/PAGE followed by autoradiography. In the cases of hormone studies, 10 nM of R5020 or 10 nM of RU486 was added during all incubation and washing steps.
Cell Culture and Transient Transfection Assays.
HeLa (human epithelial cervix carcinoma) cells were maintained in DMEM supplemented with 10% fetal bovine serum. Twenty-four hours before transfection, 105 cells were plated out per well in 12-well dishes in DMEM containing 5% dextran-coated charcoal-stripped serum. Cells were transfected with the indicated DNAs using lipofectin (Life Technologies, Grand Island, NY) according to the manufacturer’s guidelines. Twenty-four hours later, cells were washed and fed with DMEM, 5% stripped serum, and the indicated hormones. Cells were harvested 24 hr thereafter. Cell extracts were assayed for luciferase activity using the luciferase assay system (Promega), and values were corrected for protein concentration. Data are presented as the mean (±SD) of triplicate values obtained from a representative experiment that was independently repeated at least three times.
RESULTS
Effect of a Dominant-Negative Region of SRC-1A on PR Transactivation in Vitro.
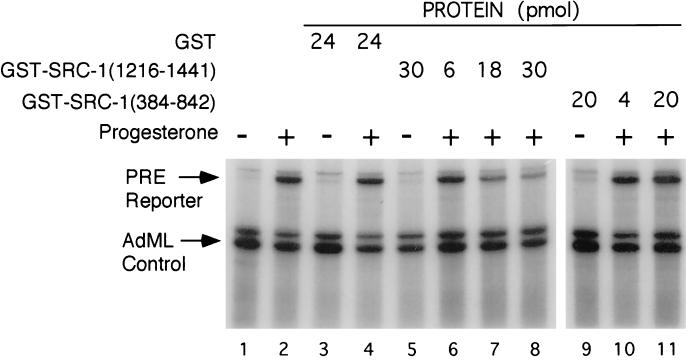
We have shown previously that the PR strongly activates transcription in vitro in the absence of any nucleosomal structure (39–41). To determine the necessity of coactivators in PR-driven transcription, we used a hormone-dependent in vitro transcription assay composed of T47D nuclear extract and a (PRE)2TATA minimal promoter linked to a G-free cassette (Fig. 1). The lengths of correctly initiated RNA transcripts from the (PRE)2TATA reporter are 273 nucleotides (upper arrow) and of read-through transcripts are 290 nucleotides. As an internal control, the AdML promoter was used, which constitutively drives transcription from a 200-bp G-free cassette and results in the synthesis of a 190-nucleotide correctly initiated RNA (lower arrow) template. As expected, addition of the PR agonist R5020 strongly activated transcription of the PRE-containing template but had no effect on the control AdML promoter template (Fig. 1, lanes 1 and 2).
Figure 1.
Role of SRC-1A in PR transactivation in vitro. Effects of purified GST control, GST-SRC-1(1216–1441), and GST-SRC-1(384–842) proteins were analyzed in a PR-driven in vitro transcription assay. Assays were performed in the presence of vehicle (lanes 1, 3, 5, and 9) or 10−7 M progesterone (lanes 2, 4, 6–8, 10, and 11) and various concentrations of GST fusion proteins (presented in pmol). Upper arrow indicates correctly initiated transcripts from the (PRE)2TATA-G-free 290-bp reporter. The AdML arrow indicates correctly initiated transcripts from the AdML-G-free 200-bp internal control. Gels were quantified using a Betagen blot scanner.
To analyze the role of SRC-1A in this in vitro PR-dependent transactivation assay, a dominant negative region of SRC-1A (amino acids 1216–1441), which strongly interacts with the PR ligand-binding domain (LBD, ref. 35), was produced as a GST fusion protein in yeast. Purified GST control protein had no significant effect on levels of the uninduced and progesterone-induced transcription (lanes 3 and 4). In contrast, the GST-SRC-1A(1216–1441) protein strongly inhibited PR driven transcription up to 75% (lanes 6–8). Another region of SRC-1A (amino acids 384–842), which does not exhibit dominant-negative properties in transient transfection studies (data not shown), did not affect the PR-driven transcription in the in vitro transcription assay (lanes 9–11). These results substantiate that transcriptional activation of PR target genes is dependent on receptor, ligand, and SRC-1A. Therefore, the coactivation function of SRC-1A or a related coactivator appears to be essential for transactivation by the liganded PR in the absence of nucleosomes.
The in Vitro Interaction Between Human PR and PCAF.
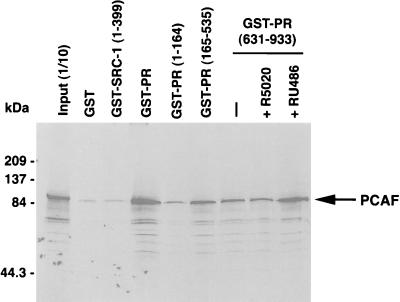
Steroid receptors can complex with CBP and SRC-1A in vitro and in vivo (33–35, 42), and each of these steroid receptor coactivators possesses intrinsic histone acetylase activity, as well as the ability to bind to each other and another HAT, PCAF (refs. 17, 34, and 43; T.E.S. et al., unpublished work). To determine if the PR also interacts with PCAF, fusion proteins of GST and various regions of the human PR were expressed in either yeast or insect cells, purified, and then tested for their ability to bind to in vitro transcribed and translated PCAF protein (Fig. 2). The GST-PRA bound PCAF, while the negative controls GST and GST-SRC1–399 did not bind well. The human PR exists in two different forms (PRB and PRA) in most tissues and cell types; the full-length human PRB consists of 933 amino acids, and amino acids 165–933 are shared by both PRB and PRA forms. As shown in Fig. 2, the first 164 amino acids that are unique to the PRB form did not bind significant amounts of PCAF. However, the common N-terminal domain (residues 165–535) and the C-terminal LBD (residues 631–933) of both PRB and PRA bound significant amounts of radiolabeled PCAF, suggesting that there are multiple PCAF interaction sites on the human PR. The PCAF interaction with the PR LBD and the full-length human PRA (data not shown) was not significantly affected by the PR agonist R5020 or the PR antagonist RU486. Thus, combined experimental results indicate that the liganded PR may recruit three histone acetyltransferases (SRC-1A, CBP, and PCAF) to target DNA promoters. This quarternary complex is likely to be very stable, because each of these proteins binds to one another. Given that each of these PR-interacting proteins possess intrinsic HAT activity, targeted histone acetylation and chromatin remodeling appears to be important to PR transactivation in vivo.
Figure 2.
Specific interaction between the human PR and PCAF in vitro. GST-hPRA, but not GST and GST-SRC1–399-bound radiolabeled PCAF. Binding PCAF was mediated by both the N terminus (GST-hPR165–535) and LBD (GST-hPR631–933). GST-PR1–164, which is hPRB specific, did not bind significant amounts of PCAF. PCAF binding to the PR LBD was not dependent on R5020 agonist or influenced by RU486 antagonist. One-tenth of the input of in vitro transcribed PCAF is shown in the first lane. Molecular mass markers are in kDa on the left.
The Histone Deacetylase Inhibitor, TSA, Potentiates Transactivation by the PR.
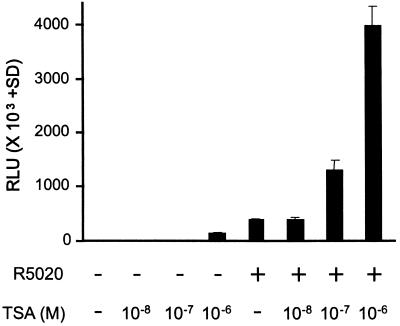
To determine effects of relative levels of histone acetylation on transactivation by the human PR, TSA, a potent inhibitor of histone deacetylases (31), was used on HeLa cells transiently transfected with wild-type hPRB and a luciferase reporter containing two PREs and a minimal TATA promoter (Fig. 3). At concentrations of 10−7 M and 10−6 M, TSA strongly potentiated PR driven transcription up to 10-fold. However, TSA also had an effect on the uninduced reporter in the absence of R5020 (Fig. 3). Given that DNA transfected by lipofectin results in formation of nucleosomal structures on episomal plasmid DNA, the effects of TSA are most likely due to inhibition of histone deacetylation. This inhibition would allow for the accumulation of acetylated histones on the promoter and the loosening of the nucleosomal structure enabling enhanced access of transcription factors. Thus, it is not surprising that both activated and unactivated promoter activity is enhanced by TSA.
Figure 3.
TSA enhances basal and PR-driven transcription in vivo. HeLa cells were transfected with 0.5 μg (PRE)2E1b-LUC reporter and 0.1 μg hPRB (DNA per three wells of a 12-well plate). Transfected cells were incubated with or without 1 nM R5020 and various concentrations (1 μM, 0.1 μM, and 0.01 μM) of TSA, a potent inhibitor of histone deacetylases. Experiments were performed in triplicate and are represented as relative light units (RLU) (±SD).
HD1 Inhibits Transactivation.
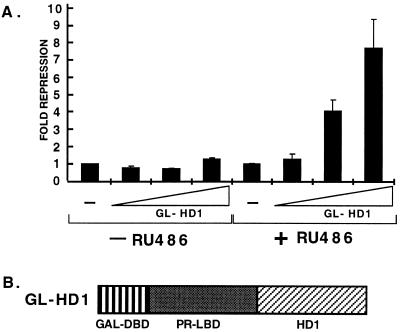
To determine the effects of histone deacetylases on PR transactivation, we used a regulatable system (37) to recruit HD1 to DNA. A hybrid fusion protein (GL-HD1) was constructed containing the GAL1–94 DNA binding domain, a C-terminal truncated human PR LBD (PRΔ42; 37), and HD1 (30). The 42-amino acid truncated PR LBD can be activated by the PR antagonist RU486, but not by PR agonist R5020 (37). The GL-HD1 was cotransfected into HeLa cells with a luciferase reporter under the control of four 17-mer GAL4 DNA binding sites (UAS)4 in front of a thymidine kinase promoter. In the absence of RU486, the GL-HD1 did not affect the basal levels of reporter activity (Fig. 4). In the presence of RU486, GL-HD1 binds DNA and represses transcription from the reporter. Increasing amounts of the transfected GL-HD1 concomitantly decreased reporter transcription. When fused to the transcriptionally active GAL1–147 DNA-binding domain, HD1 also repressed transcriptional activity of a promoter containing GAL4 binding sites (data not shown). Therefore, HD1 is a potent repressor of transcription, presumably by deacetylating histones in the promoter region and thus locking nucleosomes into a tight conformation that precludes access of transcription factors or maintains a suboptimum DNA structure.
Figure 4.
Expression of GL-HD1 inhibits transcription. (A) HeLa cells were transfected with 0.5 μg (UAS)4tk-LUC reporter, increasing amounts (0, 0.1, 0.5, and 1.0 μg) of GL-HD1 expression vector and empty vector to maintain total DNA constant at 1 μg (DNA per three wells of 6-well plate). Cells were then incubated with or without 1 nM RU486. Experiments were performed in triplicate and are represented as fold repression (±SD) as compared with the samples without GL-HD1 that were set to 1. (B) Schematic illustrating components of the GL-HD1 [GAL DNA-binding domain (DBD)1–94–PR LBDΔ42-HD1] fusion protein.
To test the ability of HD1 to affect PR transactivation further, we constructed a recombinant protein [SRC-1(1180–1441)-HD1] containing HD1 fused to a region of SRC-1A (amino acids 1180–1441), which contains a region that strongly interacts with the human PR LBD in a ligand-dependent manner (35). The SRC-1(1180–1441)-HD1 fusion protein was then cotransfected into HeLa cells along with the human PRB and a luciferase reporter driven by the (PRE)2TATA minimal promoter. The SRC-1(1180–1441)-HD1 fusion protein strongly inhibited PR-driven transcription activation in the presence of R5020 (data not shown). This effect was specific for the full-length human PR. Collectively, these results indicate that recruitment of HD1 to a liganded PR-DNA promoter complex will repress transcription and implicate roles for histone acetylation and deacetylation in controlling PR transactivation of target gene expression in vivo.
DISCUSSION
Recent discoveries have given us better insight into the mechanisms involved in gene activation by steroid receptors: (i) the cloning and characterization of coactivators that presumably bridge or enhance nuclear receptor interaction with the general transcription factors in the PIC (reviewed in refs. 10 and 11) and (ii) the finding that some of these coactivators are histone acetyltransferases and also bind other histone acetyltransferase proteins (refs. 14, 15, and 17; T.E.S. et al., unpublished work). Therefore, in vivo coactivators appear to both remodel chromatin and activate transcription; these processes are probably not mutually exclusive. Previously, it has been shown that SRC-1A and CBP synergistically enhance estrogen receptor- and PR-dependent gene transactivation (33). Here we show that PCAF also can directly interact with the PR in a ligand-independent manner. If PR, CBP, SRC-1A, and PCAF exist in one big complex on target DNA promoters, this quarternary complex should be tight and stable given that each of its components binds each of the others. It should be noted that the unliganded PR in vivo is associated with multiple heat shock proteins that may preclude the binding of PCAF (44).
The fact that the PR is able to recruit three different HAT proteins suggests that histone acetylation and chromatin remodeling is important to PR transactivation. Indeed, the histone deacetylase inhibitor TSA strongly potentiated PR transcriptional activity, suggesting that histone deacetylases exert a repressive influence on transcription. Thus, the PR may have to recruit multiple HATs, including SRC-1A, CBP, and PCAF, to counteract the repressive effects of deacetylation on promoter activity in transcriptionally repressed chromatin. Furthermore, multiple HATs may be needed to acetylate different histones in different lysines within the histone N termini or different nucleosomes surrounding the transcriptional initiation site. Gene repression and the prevention of uncontrolled transcription by tight nucleosomal structures seems to be a default pathway for most if not all genes. Therefore, the equilibrium may be biased toward deacetylation that immediately shuts down gene activation, particularly if the PIC and/or transcription activator complex becomes unstable.
In this study, recruitment of HD1 into the PR-bound complex repressed PR-driven gene activation. These results suggest that PR functions inefficiently when nucleosomes are in a closed conformation. Thus, nucleosome displacement may be an essential step toward activated gene expression. However, acetylation of histones appears not to be sufficient to enhance transcription of target genes. This is supported by observations that HAT proteins, such as PCAF and hGCN5, do not activate transcription efficiently when fused to the GAL4 DNA-binding domain (ref. 16 and unpublished observations). Another essential step in transactivation undoubtedly involves the “activation functions” of coactivators, such as SRC-1A and CBP, that do activate efficiently when fused to Gal4 DNA-binding domain, and the activation functions of the liganded steroid receptors themselves. In the present study, transcription from the (PRE)2TATA DNA template in vitro in the absence of nucleosomal structures is progesterone- and PR-dependent. A dominant-negative fragment of SRC-1A was shown to inhibit PR-driven transcription in vitro, suggesting that coactivators are essential for steroid receptor gene transactivation of target genes even in the absence of nucleosomal structure.
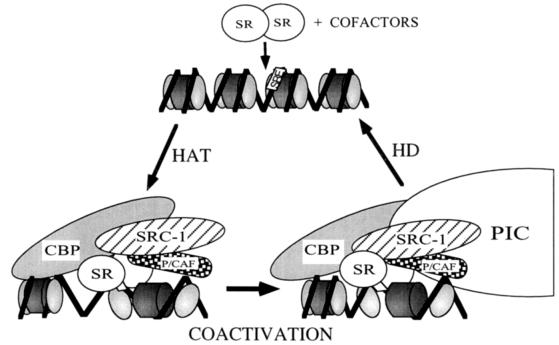
Collectively, results presented here can be used to formulate a two-step model for steroid receptor-driven transcriptional activation of target gene expression (Fig. 5). Ligand-activated steroid receptors are able to bind as dimers to cognate steroid response elements and recruit coactivators (such as CBP and SRC-1A) and PCAF into the DNA-bound complex. The accessibility of these steroid response elements within target promoters may be affected by nucleosomal structure and promoter context. The recruited HAT proteins acetylate targeted histone N termini, thereby loosening nucleosomal structure (step 1) to facilitate general transcription factor access to transcriptionally repressed chromatin. The activation functions of the coactivators, as well as the liganded steroid receptors, then enhance and stabilize assembly of the PIC (step 2). The presence of a stable steroid receptor associated complex, as well as a stable PIC, are essential, since instability could result in reversal of nucleosome displacement by histone deacetylation. Increased knowledge of the interplay of steroid receptor interaction with acetyltransferase cofactors is likely to be crucial to our understanding of how histone acetylation and deacetylation affect steroid receptor transactivation of gene expression in vivo.
Figure 5.
Two-step model of steroid receptor transactivation in vivo. Liganded steroid receptors, that are able to bind a steroid response element in chromatin repressed DNA, can recruit cofactors such as CBP, SRC-1A, and PCAF. These cofactors possess intrinsic HAT activity that can loosen nucleosomal structure by targeted histone acetylation. Coactivators, such as SRC-1A and CBP, can then initiate stable assembly of the PIC by “coactivation” that results in enhanced rates of transcription initiation by RNA polymerase II. The effects of targeted histone acetylation may be reversed by histone deacetylation (HD). Further, histone deacetylation may be responsible for turning off activated transcription once the PIC and/or steroid receptor complex become unstable.
Acknowledgments
We thank the following for materials: Y. Nakatani for pCX-Flag PCAF; S. Onate for pCBGSTPR1–164, pCBGSTPR165–535, and pCBGSTPR633–933; and S. Schreiber for pBJ-HD1-F. This work was supported by a TALENT stipend from the Netherlands Organization for Scientific Research (G.J.); National Institutes of Health National Research Service Award Postdoctoral Fellowship HD8173–01 (T.E.S.); Fritz Thyssen Foundation Postdoctoral fellowship from Germany (M.M.B.); and National Institutes of Health Grant HD08188.
ABBREVIATIONS
- PIC
preinitiation complex
- PR
progesterone receptor
- PRE
progesterone response element
- LBD
ligand-binding domain
- CREB
cAMP response element binding protein
- CBP
CREB binding protein
- AdML
adenovirus major late promoter
- GST
glutathione S-transferase
- SRC-1A
steroid receptor coactivator 1
- HAT
histone acetyltransferase
- PCAF
p300/CBP-associated factor
- HD1
histone deacetylase
- TSA
trichostatin A
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U90661).
References
- 1.Beato M. Cell. 1989;56:335–344. doi: 10.1016/0092-8674(89)90237-7. [DOI] [PubMed] [Google Scholar]
- 2.Tsai M-J, O’Malley B W. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 3.Adams C C, Workman J L. Cell. 1993;72:305–308. doi: 10.1016/0092-8674(93)90109-4. [DOI] [PubMed] [Google Scholar]
- 4.Wolffe A P. Cell. 1994;77:13–16. doi: 10.1016/0092-8674(94)90229-1. [DOI] [PubMed] [Google Scholar]
- 5.Baniahmad A, Ha I, Reinberg D, Tsai S Y, Tsai M-J, O’Malley B W. Proc Natl Acad Sci USA. 1993;90:8832–8836. doi: 10.1073/pnas.90.19.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fondell J D, Ge H, Roeder R G. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ing N H, Beekman J M, Tsai S Y, Tsai M-J, O’Malley B W. J Biol Chem. 1992;267:17617–17623. [PubMed] [Google Scholar]
- 8.Conneely O M, Kettelberger D M, Tsai M-J, O’Malley B W. In: Gene Regulation by Steroid Hormones IV. Roy A K, Clark J, editors. New York: Springer; 1989. pp. 220–223. [Google Scholar]
- 9.Tasset D, Tora L, Fromental C, Scheer E, Chambon P. Cell. 1990;62:177–1187. doi: 10.1016/0092-8674(90)90394-t. [DOI] [PubMed] [Google Scholar]
- 10.Horwitz K B, Jackson T A, Bain D L, Richer J K, Takimoto G S, Tung L. Mol Endocrinol. 1996;10:1167–1177. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- 11.Shibata H, Spencer T E, Onate S A, Jenster G, Tsai S Y, Tsai M-J, O’Malley B W. Recent Prog Horm Res. 1997;52:1–37. [PubMed] [Google Scholar]
- 12.Mitchell P J, Tjian R. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 13.Ptashne M. Nature (London) 1988;335:683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- 14.Bannister A J, Kouzarides T. Nature (London) 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 15.Ogryzko V, Schiltz L, Russanova V, Howard B H, Nakatani Y. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 16.Candau R, Moore P A, Wang L, Barlev N, Ying C Y, Rosen C A, Berger S L. Mol Cell Biol. 1996;16:593–602. doi: 10.1128/mcb.16.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. Nature (London) 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 18.Candau R, Chavez S, Beato M. J Steroid Biochem Mol Biol. 1996;57:19–31. doi: 10.1016/0960-0760(96)00262-2. [DOI] [PubMed] [Google Scholar]
- 19.Jeong S, Stein A. J Biol Chem. 1994;269:2197–2205. [PubMed] [Google Scholar]
- 20.Felsenfeld G. Proc Natl Acad Sci USA. 1996;93:9384–9388. doi: 10.1073/pnas.93.18.9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roth S Y, Allis D. Cell. 1996;87:5–8. doi: 10.1016/s0092-8674(00)81316-1. [DOI] [PubMed] [Google Scholar]
- 22.Jeong S, Stein A. Nucleic Acids Res. 1994;22:370–375. doi: 10.1093/nar/22.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Svaren J, Horz W. Genes Dev. 1996;6:164–170. doi: 10.1016/s0959-437x(96)80046-3. [DOI] [PubMed] [Google Scholar]
- 24.Archer T K, Lefebvre P, Wolford R G, Hager G L. Science. 1992;255:1573–1576. doi: 10.1126/science.1347958. [DOI] [PubMed] [Google Scholar]
- 25.Truss M, Chalepakis G, Beato M. J Steroid Biochem Mol Biol. 1992;43:365–378. doi: 10.1016/0960-0760(92)90071-p. [DOI] [PubMed] [Google Scholar]
- 26.Wolffe A P. Science. 1996;272:371–372. doi: 10.1126/science.272.5260.371. [DOI] [PubMed] [Google Scholar]
- 27.Wong J, Shi Y-B, Wolffe A P. EMBO J. 1997;11:3158–3171. doi: 10.1093/emboj/16.11.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wade P A, Wolffe A P. Curr Biol. 1997;7:82–91. doi: 10.1016/s0960-9822(06)00042-x. [DOI] [PubMed] [Google Scholar]
- 29.Wolffe A P, Pruss D. Cell. 1996;84:817–819. doi: 10.1016/s0092-8674(00)81059-4. [DOI] [PubMed] [Google Scholar]
- 30.Taunton J, Hassig C A, Schreiber S L. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 31.Yoshida M, Kijima M, Akita M, Beppu T. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 32.Vegeto E, Allan G F, Schrader W T, Tsai M-J, McDonnell D P, O’Malley B W. Cell. 1992;69:703–713. doi: 10.1016/0092-8674(92)90234-4. [DOI] [PubMed] [Google Scholar]
- 33.Smith C L, Onate S A, Tsai M-J, O’Malley B W. Proc Natl Acad Sci USA. 1996;93:8884–8888. doi: 10.1073/pnas.93.17.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 35.Onate S A, Tsai S Y, Tsai M-J, O’Malley B W. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 36.Baniahmad A, Kohne A C, Renkawitz R. EMBO J. 1992;11:1015–1023. doi: 10.1002/j.1460-2075.1992.tb05140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, O’Malley B W, Jr, Tsai S Y, O’Malley B W. Proc Natl Acad Sci USA. 1994;91:8180–8184. doi: 10.1073/pnas.91.17.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mak P, McDonnell D P, Weigel N L, Schrader W T, O’Malley B W. J Biol Chem. 1989;264:21613–21618. [PubMed] [Google Scholar]
- 39.Klein-Hitpass L, Tsai S Y, Weigel N L, Allan G F, Riley D, Rodriguez R, Schrader W T, Tsai M-J, O’Malley B W. Cell. 1990;60:247–257. doi: 10.1016/0092-8674(90)90740-6. [DOI] [PubMed] [Google Scholar]
- 40.Elliston J F, Beekman J M, Tsai S Y, O’Malley B W, Tsai M-J. J Biol Chem. 1992;267:5193–5198. [PubMed] [Google Scholar]
- 41.Bagchi M K, Tsai S Y, Tsai M-J, O’Malley B W. Nature (London) 1990;345:547–550. doi: 10.1038/345547a0. [DOI] [PubMed] [Google Scholar]
- 42.Hanstein B, Eckner R, DeRenzo J, Halachmi S, Liu H, Searcy B, Kurokawa R, Brown M. Proc Natl Acad Sci USA. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao T P, Ku G, Zhou N, Scully R, Livingston D M. Proc Natl Acad Sci USA. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith D F, Toft D O. Mol Endocrinol. 1993;7:4–11. doi: 10.1210/mend.7.1.8446107. [DOI] [PubMed] [Google Scholar]