Abstract
An earlier report showed that a disabled mutant lacking both copies of the major regulatory gene (α4) of herpes simplex virus 1 induced DNA degradation characteristic of apoptosis in infected cells, whereas the wild-type virus protected cells from apoptosis induced by thermal shock. More extensive analyses of the disabled mutant revealed a second mutation which disabled US3, a viral gene encoding a protein kinase known to phosphorylate serine/threonine within a specific arginine-rich consensus sequence. Analyses of cells infected with a viral mutant carrying a wild-type α4 gene but from which the US3 gene had been deleted showed that it induced fragmentation of cellular DNA, whereas a recombinant virus in which the deleted sequences of the US3 gene had been restored did not cause the cellular DNA to fragment. These results point to the protein kinase encoded by the US3 gene as the principal viral product required to block apoptosis.
Keywords: DNA degradation, recombinant viruses
An earlier report from this laboratory showed that cells infected with a herpes simplex virus 1 (HSV-1) mutant lacking the major regulatory gene α4 underwent apoptosis characterized by chromatin condensation, DNA degradation, etc., whereas in cells infected with wild-type virus apoptosis did not ensue (1). We also reported that wild-type virus blocked apoptosis induced by thermal shock (1). More recent studies by Koyama and Miwa (2) and also in this laboratory (V. Galvan-Girado, R.L., and B.R., unpublished results) demonstrated that the virus can also block apoptosis induced by osmotic shock. The studies carried out in connection with these experiments suggested that a functional α4 gene was necessary to block apoptosis but did not address the question of sufficiency. In an attempt to address this question, we designed several experiments that included the rescue of the deleted α4 genes in the mutant virus. To our surprise we discovered that the mutant lacking the α4 gene, HSV-1(KOS)d120 (3) had a secondary mutation in the gene US3 specifying a protein kinase (4–6). A functional US3 was required to block apoptosis.
Relevant to this report are the following:
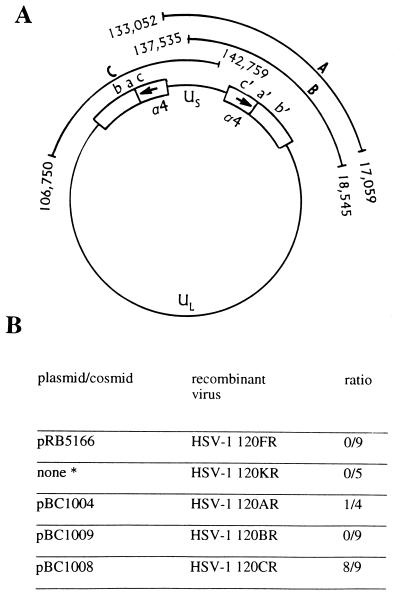
(i) The HSV-1 genome consists of two covalently linked unique sequences, a unique long sequence (UL) and a unique short sequence (US) flanked by inverted repeats (7, 8). The inverted repeats flanking UL are ab and b′a′, whereas the inverted repeats flanking US are a′c′ and ca. Upon circularization of the HSV genome the inverted repeat sequences cab and b′a′c′ separate UL from US (Fig. 1). The α4 gene maps in the c and c′ repeat sequences and therefore the viral genome contains two copies of the gene.
Figure 1.
Map of HSV-1 genome and summary of results. (A) The HSV-1 genome is represented in its circular form. Cosmids pBC1004, pBC1009, and pBC1008 are indicated as A, B, and C, respectively. The termini of the cosmid sequences carry a nucleotide number according to McGeoch et al. (9, 10). (B) Recombinant viruses were obtained by homologous recombination with either plasmid or cosmid DNA. The ∗ denotes a virus that was generated by recombination with the α4 gene sequence resident in the E5 cell line. The results are expressed as the ratio of viral plaque isolates that protected from virus-induced apoptosis to total isolates tested.
(ii) The product of the α4 gene, the infected cell protein no. 4 (ICP4), is a 1,298-amino acid multifunctional protein (8). The protein binds to viral DNA at both high- and low-affinity sites and acts as a transactivator and a repressor of viral gene function (11–19). The repressor function is associated with the presence of high-affinity sites located at or near the transcription initiation sites of several genes. The transactivating function has not been associated to specific binding to DNA, although it has been shown that late viral gene expression requires the association of ICP4 with nascent viral DNA and several host proteins (e.g., RNA polymerase II, Epstein–Barr virus small nuclear RNA-associated host protein) and ICP22, a viral transcriptional factor (20, 21). Extensive studies with deletion mutants exemplified by HSV-1(KOS)d120 led DeLuca and associates to map distinct domains within the ICP4 protein (3, 22). Studies in this laboratory have shown that ICP4 is extensively modified posttranslationally by phosphorylation, ADP-ribosylation, and nucleotidylylation (23, 24). The expectation is that these modifications are not random effects of existing enzymes but that each posttranslational modification enhances or inactivates a specific function of the protein.
(iii) Cells infected with mutants from which the α4 gene had been deleted express only the α genes—that is, the set of genes whose expression does not require prior viral DNA synthesis (3). Finally, the ICP4 in HSV-1 isolates with a limited history of passages outside the human host tends to be temperature sensitive. Cells infected and maintained at the nonpermissive temperature express primarily α protein (25). Cells maintained at 39.5°C after infection with the limited passage strain HSV-1(F) did not undergo apoptosis, in contrast with apoptosis induced in a replicate culture similarly maintained after infection with the α4 deletion mutant HSV-1(KOS)d120. This led us to conclude that α4 was required to block apoptosis (1).
(iv) HSV encodes two protein kinases expressed by the genes US3 and UL13, respectively (4, 8, 28). Whereas UL13 is packaged in the virion, US3 is not. Not all substrates of the US3 are known. The most extensively studied substrate of US3 protein kinase is an essential membrane protein encoded by the UL34 gene (5). The UL34 protein is not phosphorylated in cells infected with mutant HSV-1 lacking the US3 kinase, but in immunoprecipitation experiments the unphosphorylated UL34 protein coprecipitated with several phosphoproteins ranging in size from Mr 25,000 to 35,000 (6).
MATERIALS AND METHODS
Cells and Viruses.
Vero, HEp-2, and BHK cells were originally obtained from the American Type Culture Collection. HSV-1(F) is the prototype HSV-1 strain used in this laboratory (25). In recombinant R325 derived from HSV-1(F), approximately 800 bp constituting the carboxyl-terminal half of the α22 gene were deleted (26). d120 derived from HSV-1(KOS) contains a deletion in both copies of the α4 gene and grows only in a Vero cell line expressing the α4 gene (3). Both the virus and the cell line were kind gifts of Neal DeLuca (University of Pittsburgh). The US3 gene was deleted from the recombinant R7041 and then repaired to yield R7306 (4).
Plasmids and Cosmids.
The plasmid pRB5166 was constructed by inserting into the SalI/BamHI sites of the pACYC184 vector an HSV-1 DNA fragment that extends from the SalI site at +177 with respect to the α4 gene transcription start site and includes the entire BamHI P and BamHI S fragments. The cosmid cloning vector pRB78 was constructed as follows. The multiple cloning site of the Stratagene Supercos1 (catalog no. 251301) was cleaved with EcoRI and replaced with oligonucleotides containing cloning sites as follows: EcoRI, PacI, Sse8387I, SpeI, BamHI, NdeI, EcoRV, PacI, and EcoRI. The PacI restriction site, absent in HSV-1, serves to liberate the cloned HSV-1(F) DNA fragments from the vector. HSV-1(F) viral DNA was prepared from virions as previously described (27). To construct cosmid pBC1004, which contains the HSV-1(F) sequence nucleotide 133052 through nucleotide 17059, viral DNA was partially digested with Sau3AI, dephosphorylated, and ligated into the BamHI site of the pRB78 cosmid vector previously linearized with XbaI. The DNA was then packaged using Stratagene Gigapack XLII, following the supplier’s instructions. Escherichia coli XL-1 Blue MR was then infected, and ampicillin-resistant colonies were screened by restriction enzyme analysis. Insert termini were sequenced to verify mapping. Cosmid pBC1008 was constructed by cloning the BglII F-H fragment [HSV-1(F) nucleotides 106750 through 142759] into the BamHI site of pRB78 vector. Cosmid pBC1009 was constructed as follows. A NsiI/ScaI DNA fragment was isolated from a double digest of viral DNA. The fragment [HSV-1(F) nucleotides 137538 through 18545] was ligated into the Sse8387I/EcoRV sites of pBR78. Cosmids PBC1008 and PBC1009 were mapped by restriction enzyme analysis and insert termini were sequenced.
Construction of Recombinant Viruses by Marker Rescue.
Recombinant viruses were constructed using a modification of the technique originally described by Post and Roizman (26). Vero cells were transfected with plasmid or cosmid DNA using Lipofectamine (Life Technologies, Gaithersburg, MD), according to the supplier’s instructions. At 6 hr after transfection, the cells were exposed to 0.1–1 plaque-forming unit (pfu) of HSV-1(Kos)d120 per cell. Recombinant viruses were isolated from single plaques and grown in Vero cells. To obtain HSV-1 120KR, Vero cells were exposed to 10 pfu of HSV-1(Kos)d120 per cell. The infected cells were harvested at 48 hr after infection, frozen-thawed and sonicated, and then serially diluted and titered on Vero cells. Recombinant viruses were isolated from single plaques and grown in Vero cells.
RESULTS
HSV-1(Kos)d120 Carries an Additional Mutation.
The results described in this section emerged from studies designed to repair the deletion in both copies of the α4 gene of HSV-1(Kos)d120. We isolated recombinant viruses in which this gene was repaired by two different procedures. In the first we cloned an HSV-1(F) DNA fragment that contains an α4 sequence plus enough flanking sequence to allow homologous recombination. In this series of experiments Vero cells were transfected with plasmid DNA and infected with 0.1–1 pfu of HSV-1(Kos)d120 per cell. Under these conditions, plaques formed only in cultures of cells transfected with the plasmid DNA. Recombinant virus was recovered from individual plaques and was designated 120FR [for HSV-1(F) repair].
The second procedure was based on the observation that a small amount of virus recombines with the resident α4 gene in the E5 cell line to yield rescued virus capable of replicating efficiently in the absence of an exogenous source of ICP4. The observed recombination frequency is 10−6 to 10−7 (N. DeLuca, personal communication), and therefore such rescued virus would be expected to be present in HSV-1(Kos)d120 stock. To isolate these recombinants, Vero cells were infected at a high multiplicity with HSV-1 d120. At 24 hr after infection cells were harvested and frozen-thawed, and serial dilutions were used to infect Vero cells. The recombinant virus obtained in this fashion was designated 120KR [for HSV-1(KOS) repair].
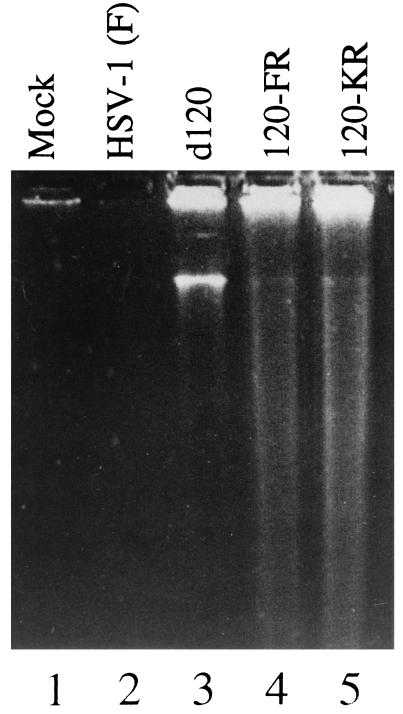
As shown in Fig. 2, analyses of DNA extracted from replicate Vero cell cultures infected with HSV-1(KOS)d120, 120FR, or 120KR showed ladders typical of apoptotic cells. These ladders were absent from extracts of mock-infected cells or cells infected with HSV-1(F). We conclude from these experiments that HSV-1(KOS)d120 genome contains an additional mutation other than in the α4 gene.
Figure 2.
Photograph of an agarose gel containing electrophoretically separated DNA fragments and stained with ethidium bromide. Vero cells were mock-infected (lane 1) or infected with HSV-1(F) (lane 2), HSV-1 d120 mutant (lane 3), HSV-1 120FR mutant (lane 4), or HSV-1 120KR mutant (lane 5). At 30 hr after infection, 2 × 106 cells per sample were collected, rinsed with PBS, lysed in a solution containing 10 mM Tris⋅HCl at pH 8.0, 10 mM EDTA, and 0.5% Triton X-100, and centrifuged at 12,000 rpm for 25 min in an Eppendorf microcentrifuge to pellet chromosomal DNA. Supernatant fluids were digested with 0.1 mg of RNase A per ml at 37°C for 1 hr and then for 2 hr with 1 mg of proteinase K per ml at 50°C in the presence of 1% sodium dodecyl sulfate (SDS), extracted with phenol and chloroform, and precipitated in cold ethanol and subjected to electrophoresis on horizontal 1.5% agarose gels containing 5 μg of ethidium bromide per ml. DNA was visualized by UV light transillumination. Photographs were taken with the aid of a computer-assisted image processor (Eagle Eye II; Stratagene).
The US3 Protein Kinase Is Required to Block Apoptosis Induced by HSV-1 Infection.
In the following series of experiments we defined the region that contains the additional mutation in HSV-1(KOS)d120 by rescue of the HSV-1(KOS)d120 with cosmids containing large HSV-1 DNA fragments. The three cosmids used in these studies were cosmid pBC1008, which contains all of the HSV-1 terminal repeat sequence, almost all of the US sequence, and part of the UL sequence (Fig. 1), and cosmids pBC1004 and pBC1009, which contain fragments spanning the entire repeat sequence but differ in the extent of coverage of the US region. Individual plaque-purified isolates from each transfection were tested for their ability to protect infected cells from apoptosis induced by the infection. The results shown in Fig. 1B suggested that the second mutation in HSV-1(KOS)d120 may map in the US domain containing the genes US1 through US3.
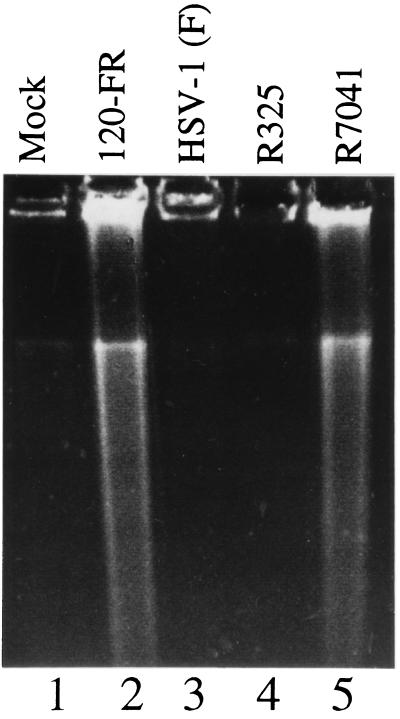
To map the function required to block apoptosis more precisely, we took advantage of the availability in this laboratory of several deletion mutants which span the sequence thought to encode the gene required to block apoptosis in infected cells. Recombinant virus R325 lacks the carboxyl-terminal half of the α22 gene, virtually all of the US1.5 gene, which overlaps the carboxyl half of the α22 gene, and the 3′ domain of the US2 gene. Recombinant virus R7041 lacks most of the US3 gene, whereas in recombinant R7306 the US3 gene had been repaired. The results, shown in Fig. 3, indicate that apoptosis was induced in cells infected with R7041 (ΔUS3) but not in cells infected with the other mutants.
Figure 3.
Photograph of an agarose gel containing electrophoretically separated DNA fragments and stained with ethidium bromide. Vero cells were mock-infected (lane 1), infected with HSV-1 120FR mutant (lane 2), wild-type HSV-1(F) (lane 3), HSV-1 R325 mutant (lane 4), or HSV-1 R7041 mutant (lane 5). Cells were harvested and processed as described in the legend to Fig. 2.
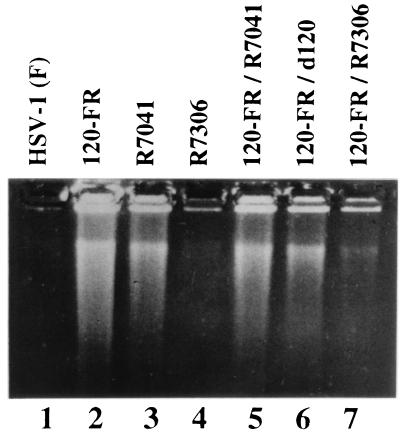
Two series of experiments verified the conclusion that a functional US3 gene is required for prevention of apoptosis and that the second mutation in the HSV-1(KOS)d120 resides in the US3 gene. In the first series we carried out simple complementation tests to determine whether the second mutation in HSV-1(KOS)d120 was the same as that in the rescued virus or in R7041 (ΔUS3) recombinant. Specifically, Vero cells were infected with artificial mixtures of 120FR and HSV-1(KOS)d120, 120FR and R7041, or 120FR and R7306 (US3 repaired). The results, shown in Fig. 4, indicated the following. As could be expected, 120FR did not complement HSV-1(KOS)d120, the parent virus from which it was derived. 120FR and R7041 also did not complement each other, suggesting that 120FR and its parent virus, HSV-1(KOS)d120 contained a nonfunctional US3. Verification of this hypothesis emerged from the observation that R7306 containing a repaired US3 gene complemented 120FR and blocked apoptosis in cells infected with these two viruses.
Figure 4.
Photograph of an agarose gel containing electrophoretically separated DNA fragments and stained with ethidium bromide. Vero cells were infected with HSV-1(F) (lane 1), HSV-1 120FR mutant (lane 2), HSV-1 R7041 mutant (lane 3), HSV-1 R7306 mutant (lane 4), or double-infected with HSV-1 120FR and R7041 mutants (lane 5), HSV-1 d120 and 120FR mutants (lane 6), or HSV-1 120FR and R7306 mutants (lane 7). Cells were harvested and processed as described in the legend to Fig. 2.
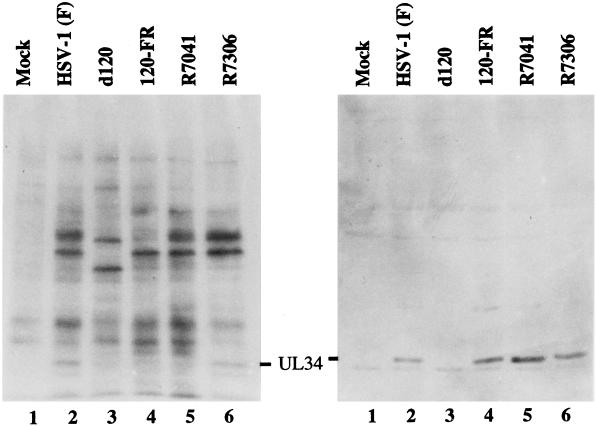
The second series of experiments served to verify that HSV-1(KOS)d120 and its derivatives lacked a functional US3 gene. As described in the Introduction, the product of UL34 gene has been shown in earlier studies from this laboratory to be a substrate for the US3 protein kinase. In wild-type-infected cells, UL34 is phosphorylated and has an apparent Mr of 30,000. Inasmuch as the original experiments were done in BHK cells, in this series of experiments replicate cultures of BHK cells were infected with 10 pfu of HSV-1(F), 120FR, R7041, or R7306 per cell. The results, shown in Fig. 5, indicate that the fully processed UL34 protein was present in cells infected with HSV-1(F) but not cells infected with 120FR or R7041. As could be expected, the cells infected with HSV-1(KOS)d120 did not make detectable quantities of UL34 protein inasmuch as they lack the α4 gene. Taken together, our results indicate that the US3 kinase is functionally absent from HSV-1(KOS)d120 and 120FR recombinant viruses. We conclude that US3 is required for protection from apoptosis induced by HSV-1.
Figure 5.
BHK-C13 cells were infected with 10 pfu of indicated viruses per cell. At 13 hr after infection the cells were incubated in phosphate-free medium for 1 hr and then labeled in the same medium supplemented with 32Pi for 4 hr as described earlier (5). Cell lysates were electrophoretically separated in SDS/polyacrylamide gels and electrically transferred onto nitrocellulose filters. Filters were exposed to film or allowed to react with rabbit polyclonal antibody to the UL34 protein as described earlier (5).
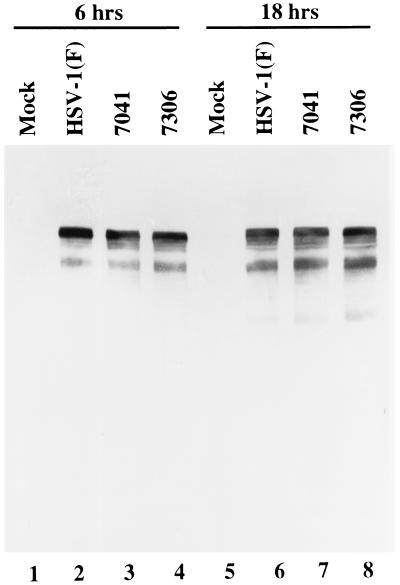
Last, it was of interest to determine whether US3 phosphorylates ICP4 in infected cells. The US3 protein kinase phosphorylates threonine/serine in the consensus sequence RRR-R/X-S/T-R/Y where Y is defined as any amino acid with the exception of acid residues and a preference for Arg, Ala, Val, or Ser (28, 29). Although ICP4 lacks a consensus site for phosphorylation by US3, we compared the electrophoretic mobility of ICP4 from cells infected with wild type, mutant R7041 (ΔUS3), or a recombinant virus R7306 in which the deleted UL3 sequence had been restored. ICP4 forms several bands differing in electrophoretic mobility in denaturing gels, and in some instances the lack of a specific band correlated with the absence of a specific posttranslational modification by a viral kinase (31). The results of the assay shown in Fig. 6 are not consistent with the hypothesis that the posttranslational modifications of ICP4 are due to phosphorylation by the US3 kinase.
Figure 6.
Photograph of electrophoretically separated proteins from HEp-2 cells either mock-infected or infected with 10 pfu of indicated virus per cell and harvested 6 or 18 hr after infection. A total of 2 × 105 cells per sample were disrupted in SDS and 2-mercaptoethanol-containing buffer, boiled for 3 min, and electrophoretically separated on an SDS/N,N′-diallyltartardiamide/9.5% polyacrylamide gel at 4°C. The denatured, electrophoretically separated polypeptides were electrically transferred to a nitrocellulose sheet, where they were allowed to react with an anti-ICP4 monoclonal antibody (H640).
DISCUSSION
The key discovery reported here is that a functional protein kinase encoded by the US3 gene of herpes simplex virus is required to block apoptosis induced by infection and also, as previously described, by thermal shock. This discovery raises several issues that require a detailed exposition.
(i) Genetically engineered mutants produced by recombination following transfection frequently acquire secondary mutations that may not be readily detectable unless specifically looked for. As a general rule, this laboratory rescues genetically engineered mutations to ensure that the phenotype can be directly related to the specific mutation. In this instance the mutation in US3 was silent and would not have affected the studies carried out by N. DeLuca and associates, since viral gene expression, including that of US3, requires a functional α4 gene and apoptosis is a very late event. Even if a rescue had been done, the mutation in US3 would not have been detected unless the infected cell had been tested for the US3 function. In a different vein, the block of apoptosis in cells infected with HSV-1(F) and maintained at 39.5°C suggests that the temperature-sensitive mutation may not have been very tight and that amounts of US3 protein sufficient to block apoptosis were made.
(ii) The US3 protein kinase phosphorylates threonine/serine in the consensus sequence RRR-R/X-S/T-R/Y (28, 29). In the case of UL34, this was verified by mutagenesis of the threonine codon in the sequence encoding RRRRTRRSRE (6). It could be predicted that US3 blocks apoptosis by phosphorylating one or more proteins, but their identity is currently unknown. ICP4 does not appear to be a substrate of US3 protein kinase, but we have not totally excluded the involvement of this protein in blocking apoptosis.
(iii) US3 is dispensable for viral growth in cell culture (4). It is of interest, however, that cells infected with this mutant have a crenated appearance expected from cells undergoing osmotic shock. Although virus yield is not grossly affected in cells in culture, R7041 recombinant is virtually apathogenic on intracerebral inoculation in mice [1.8 × 106 pfu/LD50, compared with 102 pfu/LD50 for the parent, HSV-1(F) virus in the same assay] (30). One explanation for the absence of a significant effect on R7041 virus yield in cell culture is that in the systems examined to date the manifestation of apoptosis—i.e., degradation of host DNA—occurs late in infection, virtually after the completion of the reproductive cycle.
The involvement of US3 protein kinase in blocking apoptosis induced by infection or thermal or osmotic shocks suggests that HSV differs from other viruses in the mechanism by which it blocks the apoptosis as a host response to infection. It raises the possibility that US3 may find utility in controlling apoptosis independent of viral infection.
Acknowledgments
We thank Neal DeLuca for the HSV-1 mutant d120 and the ICP4-expressing cell line E5. These studies were aided by grants from the National Cancer Institute (CA47451), the U.S. Public Health Service.
ABBREVIATIONS
- HSV-1
herpes simplex virus 1
- UL
unique long sequence
- US
unique short sequence
- ICPn
infected cell protein no. n
- pfu
plaque-forming unit
References
- 1.Leopardi R, Roizman B. Proc Natl Acad Sci USA. 1996;93:9583–9587. doi: 10.1073/pnas.93.18.9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koyama A H, Miwa Y. J Virol. 1997;71:2567–2571. doi: 10.1128/jvi.71.3.2567-2571.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeLuca N A, McCarthy A, Schaffer P A. J Virol. 1985;56:558–570. doi: 10.1128/jvi.56.2.558-570.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Purves F C, Longnecker R M, Leader D P, Roizman B. J Virol. 1987;61:2896–2901. doi: 10.1128/jvi.61.9.2896-2901.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Purves F C, Spector D, Roizman B. J Virol. 1991;65:5757–5764. doi: 10.1128/jvi.65.11.5757-5764.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Purves F C, Spector D, Roizman B. J Virol. 1992;66:4295–4303. doi: 10.1128/jvi.66.7.4295-4303.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wadsworth S, Jacob R J, Roizman B. J Virol. 1975;15:1487–1497. doi: 10.1128/jvi.15.6.1487-1497.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roizman B, Sears A E. In: Fields’ Virology. 3rd Ed. Fields B N, Knipe D M, Howley P, Chanock R M, Hirsch M S, Melnick J L, Monath T P, Roizman B, editors. New York: Raven; 1995. pp. 2231–2295. [Google Scholar]
- 9.McGeoch D J, Dolan A, Donald S, Rixon F J. J Mol Biol. 1985;181:1–13. doi: 10.1016/0022-2836(85)90320-1. [DOI] [PubMed] [Google Scholar]
- 10.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 11.Preston C M. J Virol. 1979;29:275–284. doi: 10.1128/jvi.29.1.275-284.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kristie T M, Roizman B. Proc Natl Acad Sci USA. 1986;83:3218–3222. doi: 10.1073/pnas.83.10.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kristie T M, Roizman B. Proc Natl Acad Sci USA. 1986;83:4700–4704. doi: 10.1073/pnas.83.13.4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faber S W, Wilcox K W. Nucleic Acids Res. 1986;14:6067–6083. doi: 10.1093/nar/14.15.6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muller M T. J Virol. 1987;61:858–865. doi: 10.1128/jvi.61.3.858-865.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michael N, Spector D, Mavromara-Nazos P, Kristie T M, Roizman B. Science. 1988;239:1531–1534. doi: 10.1126/science.2832940. [DOI] [PubMed] [Google Scholar]
- 17.Michael N, Roizman B. Proc Natl Acad Sci USA. 1989;86:9808–9812. doi: 10.1073/pnas.86.24.9808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michael N, Roizman B. Proc Natl Acad Sci USA. 1993;90:2286–2290. doi: 10.1073/pnas.90.6.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leopardi R, Michael N, Roizman B. J Virol. 1995;69:3042–3048. doi: 10.1128/jvi.69.5.3042-3048.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leopardi R, Roizman B. Proc Natl Acad Sci USA. 1996;93:4572–4576. doi: 10.1073/pnas.93.10.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leopardi R, Ward P, Ogle W, Roizman B. J Virol. 1997;71:1133–1139. doi: 10.1128/jvi.71.2.1133-1139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeLuca N A, Schaffer P A. J Virol. 1988;62:732–743. doi: 10.1128/jvi.62.3.732-743.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blaho J A, Michael N, Kang V, Aboul-Ela N, Smulson M E, Jacobson M K, Roizman B. J Virol. 1992;66:6398–6407. doi: 10.1128/jvi.66.11.6398-6407.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blaho J A, Roizman B. J Virol. 1991;65:3759–3769. doi: 10.1128/jvi.65.7.3759-3769.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ejercito P M, Kieff E D, Roizman B. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 26.Post L E, Roizman B. Cell. 1981;25:227–232. doi: 10.1016/0092-8674(81)90247-6. [DOI] [PubMed] [Google Scholar]
- 27.Kieff E D, Bachenheimer S L, Roizman B. J Virol. 1971;8:125–132. doi: 10.1128/jvi.8.2.125-132.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Purves F C, Donella-Deana A, Marchiori F, Leader D P, Pinna L A. Biochim Biophys Acta. 1986;889:208–215. doi: 10.1016/0167-4889(86)90106-0. [DOI] [PubMed] [Google Scholar]
- 29.Leader D P, Donella-Deana A, Marchiori F, Purves S P, Pinna L A. Biochim Biophys Acta. 1991;1091:426–431. doi: 10.1016/0167-4889(91)90210-o. [DOI] [PubMed] [Google Scholar]
- 30.Meignier B, Longnecker R, Mavromara-Nazos P, Sears A E, Roizman B. Virology. 1988;162:251–254. doi: 10.1016/0042-6822(88)90417-5. [DOI] [PubMed] [Google Scholar]
- 31.Purves F C, Ogle W O, Roizman B. Proc Natl Acad Sci USA. 1993;90:6701–6705. doi: 10.1073/pnas.90.14.6701. [DOI] [PMC free article] [PubMed] [Google Scholar]