Abstract
Polyadenylation of premessenger RNAs occurs posttranscriptionally in the nucleus of eukaryotic cells by cleavage of the precursor and polymerization of adenosine residues. In the yeast Saccharomyces cerevisiae, the mature poly(A) tail ranges from 60 to 70 nucleotides. 3′-end processing can be reproduced in vitro with purified factors. The cleavage reaction requires cleavage factors I and II (CF I and CF II), whereas polyadenylation involves CF I, polyadenylation factor I (PFI), and poly(A) polymerase (Pap1p). CF I has recently been separated into two factors, CF IA and CF IB. We have independently purified CF IA and found that five polypeptides cofractionate with the activity. They include Rna14p, Rna15p, Pcf11p, a new protein called Clp1p, and remarkably, the major poly(A)-binding protein Pab1p. Extracts from strains where the PAB1 gene is mutated or deleted are active for cleavage but generate transcripts bearing abnormally long poly(A) tracts. Complementation with recombinant Pab1p not only restores the length of the poly(A) tails to normal, but also triggers a poly(A) shortening activity. In addition, a monoclonal Pab1p antibody prevents the formation of poly(A) tails in extracts or in a reconstituted system. Our data support the notion that Pab1p is involved in the length control of the poly(A) tails of yeast mRNAs and define a new essential function for Pab1p in the formation of mature mRNAs.
The formation of poly(A) tails at the 3′ end of premessenger RNAs is an obligatory step in the maturation of most eukaryotic transcripts. In the yeast Saccharomyces cerevisiae, as in mammalian cells, 3′-end processing occurs by cleavage of the precursor RNA, generating the upstream cleavage product that is elongated by addition of a polyadenylate tail (1–3). Cis-acting elements (4) as well as trans-acting proteins are essential for 3′ processing. While tightly coupled in vivo, the cleavage and polyadenylation reactions can be studied separately in vitro with either whole cell extracts (4–6) or with purified factors. Chen and Moore (7) reported the first fractionation of yeast extracts into four chromatographic fractions that can reproduce 3′-end processing of synthetic transcripts in vitro. Cleavage factors I and II (CF I and CF II) are necessary and sufficient to cleave the mRNA precursor molecule. In addition to CF I, poly(A) polymerase (Pap1p), and polyadenylation factor I (PF I) are required to specifically polyadenylate the upstream cleavage fragment (7). The PAP1 gene product, Pap1p, was the first identified component of the 3′-end processing machinery (8, 9). Unlike its mammalian counterpart, Pap1p is not required in vitro for the cleavage reaction (7). By virtue of synthetic lethality with pap1 mutant alleles, we demonstrated that the proteins encoded by the RNA14 and RNA15 genes are involved in 3′-end processing and that they are essential components of CF I (10). Extracts prepared from either rna14 or rna15 mutant strains are inactive for both cleavage and polyadenylation (10). A recent study showed that CF I can be separated into two activities, CF IA and CF IB (11). CF IA is a tetrameric protein complex with subunits of 76, 70, 50, and 38 kDa. The 76- and 38-kDa polypeptides have been shown to be Rna14p and Rna15p, respectively, confirming our previous data. CF IB contains only one single polypeptide of 73 kDa that remains unidentified so far (11). We have independently purified CF IA by following the complementation of an rna15 mutant extract and found polypeptides with the same apparent molecular masses as those reported by Kessler et al. (11) cofractionating with the activity. Peptide microsequencing showed that the 50-kDa protein is encoded by a novel gene we termed CLP1 (cleavage/polyadenylation factor IA subunit). A detailed study of this subunit will be published separately (L.M.-S. and W.K., unpublished work). A careful analysis revealed that the band running as a 70-kDa protein was a mixture of two different polypeptides with similar apparent molecular masses. One was the recently described Pcf11 protein (12) and the other was surprisingly identified as the major poly(A)-binding protein Pab1p (13). This protein has previously been implicated in cytoplasmic events such as translation initiation (14–17), mRNA decapping (18), and deadenylation by stimulation of a poly(A) nuclease (19, 20). Here, we demonstrate the involvement of Pab1p in 3′-end formation. We examined the 3′-end processing activity of extracts where Pab1p is absent or present as a temperature-sensitive protein, or is inactivated by a specific antibody. In all cases, poly(A) tail formation is impaired. A detailed analysis showed that Pab1p is not involved in cleavage but is required for the synthesis of poly(A) tails of normal length. We propose that in yeast poly(A) length control is achieved through inhibition of poly(A) polymerase activity by Pab1p.
MATERIALS AND METHODS
Purification of CF IA.
The detailed purification of CF IA, as well as the characterization of the 50-kDa subunit (Clp1p), will be described elsewhere (L.M.-S. and W.K., unpublished work). In brief, extracts of a strain expressing a histidine-tagged version of Rna15p (LM106; Matα, ade2, leu2, ura3, trp1, his3, YCp-ADE2-[His]6-RNA15) were prepared (6). CF IA was purified over four columns to homogeneity by complementation of an rna15 mutant extract (10). The columns used were: DEAE-Sepharose (Pharmacia), hydroxyapatite, Ni2+-nitrilotriacetic-agarose (Ni2+-NTA; Qiagen, Chatsworth, CA), and poly(U)-Sepharose (Pharmacia). The CF II/CF IB fraction used in this study was from the hydroxyapatite column and is free of PF I activity.
Yeast Strains.
The wild-type YAS100 strain, the pab1-F364L temperature-sensitive mutant (YAS120), and the pab1Δ rpl46Δ strain (YAS394) have been described elsewhere (15). Strain YAS1437 (MATa, ade2, leu2, ura3, trp1, his3, pab1::HIS3, spb8-1) contains a deletion of the PAB1 gene suppressed by the spb8-1 mutation. The YAS strains were generous gifts of Alan Sachs (University of California, Berkeley).
Recombinant Proteins and Antibodies.
Recombinant Pab1p (21) was a kind gift of Alan Sachs. Polyclonal antibodies to Rna14p, Rna15p, and Clp1p were obtained by immunizing rabbits with recombinant proteins purified over a Ni2+-NTA column (Qiagen). Pab1p and Pub1p mAbs (22) were kindly provided by Maurice Swanson (University of Florida, Gainesville). Anti-Pcf11p (12) were a kind gift of François Lacroute (Centre de Génétique Moléculaire, Gif-sur-Yvette, France).
Processing Assays.
Extracts were prepared according to Butler et al. (6). Standard 3′-processing assays with CYC1 and CYC1 precleaved precursors were performed as described (10, 23). Poly(U)-Sepharose CF IA fractions (3 μl) were used in complementation assays with 45 μg of rna15 mutant extracts per reaction. In the reconstitution experiments, 2 μl of purified CF IA [poly(U)-Sepharose fraction], 2 μl of partially purified CF II/CF IB (hydroxyapatite fraction), and 0.5 μl of purified PF I/Pap1p (Mini Q fraction; ref. 24) were used. For complementation assays with Pab1p, the protein was preincubated with the extract for 15 min at room temperature. In the antibody inactivation experiments with extracts, 0.5 μl of mAb to Pab1p (22) was preincubated with 25 μl (450 μg of protein) of the wild-type YAS100 extract for 30 min. For inactivation experiments with purified factors, 10 μl of CF IA were preincubated with 0.5 μl of increasing dilutions of monoclonal Pab1p antibody (150, 75, 15, and 1.5 ng) for 15 min at room temperature. Two microliters were withdrawn, added to the rest of the reaction mixture, and the reactions were started. RNA products were analyzed on 5% polyacrylamide/8 M urea gels and visualized by autoradiography.
RESULTS
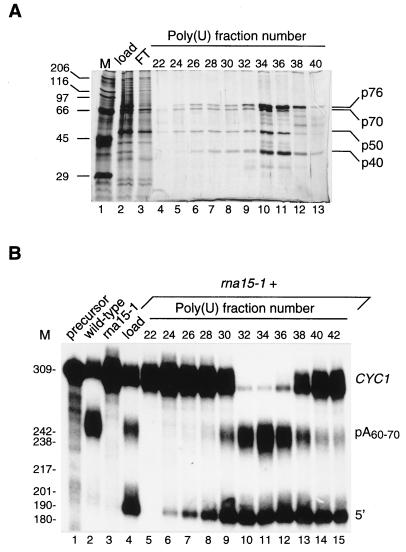
We have previously reported the characterization of Rna14 and Rna15 proteins as components of CF I, one of the four factors required for mRNA 3′-end formation in S. cerevisiae (10). A yeast strain expressing Rna15p with an amino-terminal histidine tag was constructed and used to purify the factor that complements the cleavage and polyadenylation deficiency of an rna15 mutant extract (10). Poly(U)-Sepharose affinity chromatography was the final step of the purification (see Materials and Methods). Four polypeptides of molecular masses 76, 70, 50, and 40 kDa copurified with the complementing activity (Fig. 1).
Figure 1.
Purification of CF IA by complementation of an rna15 mutant extract. (A) SDS/polyacrylamide gel of the final poly(U)-Sepharose column fractions. Trichloroacetic acid-precipitated aliquots of the load (lane 2), the flow-through (lane 3), and every second fractions from 22 to 40 (lanes 4–13) were resolved on an SDS/10% polyacrylamide gel and stained with silver. Sizes (in kDa) of protein markers (M, lane 1) are given on the left. CF IA subunits are indicated on the right. (B) Complementation of an rna15 mutant extract with the poly(U)-Sepharose fractions. Lane 1, unreacted CYC1 precursor. Lane 2, reaction with wild-type extract. Reactions of rna15 extract either alone (lane 3) or complemented with the poly(U)-Sepharose load (lane 4), or with column fractions (lanes 5–15). Sizes of DNA markers are given on the left (M; in nucleotides). Position of the precursor (CYC1), the upstream cleavage product (5′), and the polyadenylated RNA (pA60–70) are indicated on the right.
A recent study reported the separation of CF I into two activities, CF IA and CF IB, both required for cleavage and polyadenylation (11). Whereas CF IB is a unique protein of 73 kDa, CF IA contains four polypeptides the apparent molecular masses of which are in good agreement with those of our CF I components. It is very likely that in our purification, CF IB copurified with CF II because we could not reconstitute the polyadenylation reaction with a precleaved CYC1 transcript (23) with our purified CF I, PF I and Pap1p proteins unless the partially purified CF II fraction was added (data not shown). For this reason, we will refer now to our poly(U)-Sepharose fractions as CF IA.
The complete 3′-processing reaction was reconstituted with purified CF IA [poly(U) fraction 34; Fig. 1A, lane 10], partially purified CF II/CF IB (see Materials and Methods), and purified PF I/Pap1p [Mini Q fraction; (24)]. PF I, only required for polyadenylation (7), has been purified as a large complex containing multiple subunits, including poly(A) polymerase (24). This explains why this factor, referred to as PF I/Pap1p, was able to elongate nonspecifically the CYC1 precursor in a polyadenylation reaction (Fig. 2, lane 6). CF IA and CF II/CF IB together generated the expected cleavage product (Fig. 2, lane 5), but they were not active separately (see Fig. 2, lanes 3 and 4). Upon combination, CF IA, CF II/CF IB, and PF I/Pap1p gave rise to mature polyadenylated transcripts (Fig. 2, lane 7; pA60–70). The poly(A) tails have an average size of 60–70 nucleotides, which is in agreement with previous observations in vivo and in unfractionated extracts (for review, see ref. 2). This result suggests that the proteins from the poly(U)-Sepharose fraction are sufficient to act as CF IA in 3′-end formation of mRNAs.
Figure 2.
In vitro reconstitution of a 3′-end processing reaction with purified factors. Lane 1, DNA markers (in nucleotides). Lane 2, unreacted CYC1 precursor. Purified CF IA [poly(U)-Sepharose fraction 34], partially purified CF II/CF IB, and purified PF I/Pap1p were incubated with CYC1 either alone (lanes 3, 4, 6, respectively) or together (CF IA + CF II/CF IB + PF I/Pap1p, lane 7). CYC1, precursor; 5′, upstream fragment; pA60–70, pAn, polyadenylated transcripts.
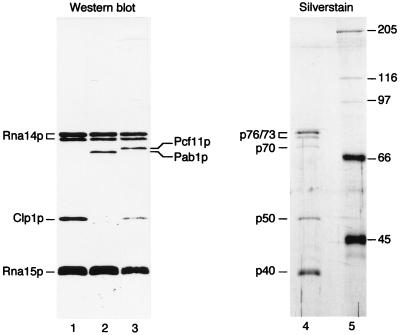
To investigate further the composition of CF IA, aliquots of the poly(U)-Sepharose fraction 35 (see Fig. 1A) were run in parallel on an SDS/8% polyacrylamide gel. One lane was stained with silver (Fig. 3, lane 4), and the other one used for a Western blot analysis (Fig. 3, lane 1–3). Probing the blot with antibodies to Rna14p identified the doublet of 76/73 kDa on the silver-stained gel (Fig. 3, lane 4) as Rna14p (Fig. 3, lane 1). Antibodies to Rna15p gave a broad 40-kDa signal (Fig. 3, lane 1) that also corresponds to a doublet that is weakly visible on the silver-stained gel (Fig. 3, lane 4). The size of Rna15p is slightly larger than reported (11) due to the presence of a histidine tag at its amino terminus. The multiple forms of Rna14p and Rna15p were found reproducibly and might result from degradation or from posttranslational modification. These data confirm our previous results (10) showing that Rna14p and Rna15p copurify with CF I activity. Microsequencing of the 50-kDa band allowed us to clone a gene that we called CLP1 (L.M.-S. and W.K., unpublished work). Antibodies raised against recombinant Clp1p recognized the 50-kDa polypeptide in CF IA (Fig. 3, lane 1).
Figure 3.
Identification of the 76/73-, 70-, 50-, and 40-kDa subunits of CF IA. Proteins from poly(U)-Sepharose fraction 35 were resolved on a 20-cm long SDS/8% polyacrylamide gels and analyzed by Western blot (lane 1–3) or silver stain (lane 4). The Western blot was probed with the following dilutions of antibodies: Anti-Rna14p (1:2500; lanes 1–3); anti-Rna15p (1:10000; lanes 1–3); anti-Clp1p (1:1000; lanes 1 and 3); anti-Pab1p (1:1000; lane 2); and anti-Pcf11p (1:5000; lane 3). Lane 5, protein markers (sizes, in kDa are given on the right).
The observation that CF IA, but not Rna15p, also binds to poly(A)-Sepharose (ref. 11, and our unpublished data), and that the 70-kDa subunit of our purified factor is reminiscent of the size of the major poly(A)-binding protein (13), prompted us to strip and reprobe the blot with a mAb to Pab1p (22) together with antibodies to Rna14p and Rna15p. The Pab1p antibody recognized a 70-kDa protein that we therefore considered as Pab1p (Fig. 3, lane 2).
Amrani et al. (12) recently reported the characterization of an essential protein called Pcf11p (protein 1 of CF I) based on interactions with Rna14p and Rna15p in the two-hybrid system; pcf11 mutant extracts are deficient in 3′-end processing. Based on these data, Amrani et al. proposed that PCF11 encodes a new component of CF I (12). Since both Pcf11p and Pab1p have an apparent molecular mass of 70 kDa, it could be that the 73-kDa polypeptide associated with CF IB in Kessler et al. (11) is Pcf11p or that the 70-kDa band of CF IA is a heterogeneous mixture of two proteins. We stripped the blot and reprobed it with antibodies to Pcf11p, Rna14p, Rna15p, and Clp1p. Antibodies to Pcf11p actually revealed a protein in CF IA (Fig. 3, lane 3). The alignment of Rna14p and Rna15p signals (Fig. 3, lanes 2 and 3) showed that Pcf11p ran slightly slower than Pab1p. It was not possible to distinguish these two proteins on the silver-stained gel. Several explanations could account for these discrepancies: (i) the silver-staining procedure does not allow a proper detection of either Pab1p or Pcf11p; and (ii) the amount of one of the polypeptides is below the threshold of detection by this technique. We will focus for the rest of this study on the copurification of Pab1p with CF IA activity.
Previous studies demonstrated that Pab1p is required for translation initiation (14, 16) and may also participate in mRNA decapping and deadenylation (18). So far, the roles assigned to Pab1p take place in the cytoplasm, in agreement with its predominant localization (22). To obtain evidence for a nuclear role of Pab1p in 3′-end formation of pre-mRNAs, we added the Pab1p mAb to wild-type extract. We could observe a lower amount of cleavage intermediate and a smear overlapping with the CYC1 precursor (Fig. 4A; compare lanes 2 and 3). In the presence of the chain terminator cordycepin triphosphate, the cleavage activity also decreased, probably reflecting a partial disruption of the cleavage complex by the mAb. In addition, the smear disappeared proving it was due to an increase of the poly(A) tail length (data not shown). The same amount of an unrelated mAb did not affect the reaction (data not shown).
Figure 4.
Pab1p is required for pre-mRNA 3′-end processing. (A) Inhibition of wild-type extract by a Pab1p mAb. The 3′-processing activity of the wild-type YAS100 extract (lane 2) is inhibited by the addition of mAb to Pab1p (22) (lane 3). Addition of increasing amounts of recombinant Pab1p (21) (200 ng, 500 ng, 1 μg, and 2 μg; lanes 4–7, respectively) restores 3′-processing. (B) Abnormal polyadenylation reaction in pab1 mutant extracts. Extracts from YAS120 (pab1-F364L), YAS394 (pab1Δ rpl46Δ), and YAS1437 (pab1Δ spb8-1) strains give products with poly(A) tails longer than normal (lanes 1, 6, and 11, respectively). Increasing amounts of recombinant Pab1p (21) (50, 100, 200, and 500 ng; lanes 2–5, 7–10, and 12–15, respectively) give rise to species with poly(A) tails of normal length. M, DNA markers. Lane 1, CYC1 precursor; 5′, upstream fragment; pA, pAn, polyadenylated transcripts.
We then prepared extracts from strains either bearing a pab1 temperature-sensitive mutation (pab1-F364L, ref. 14) or a deletion rescued by two different suppressors (pab1Δ spb8-1 or pab1Δ rpl46Δ, ref. 14). With these extracts, the upstream fragment received poly(A) tails [as confirmed by chromatography on poly(U)-Sepharose and subsequent gel electrophoresis; data not shown] longer than in wild-type extracts [compare the polyadenylated species (pA) in Fig. 4A, lane 2, with pAn in Fig. 4B, lanes 1, 6, and 11]. Adding increasing amounts of recombinant Pab1p (21) to either the immunoinactivated or the mutant pab1 extracts resulted in poly(A) tails with normal size (Fig. 4A, lanes 4–7; Fig. 4B, lanes 2–5, 7–10, and 12–15). The higher amount of Pab1p required to reconstitute the reaction likely reflects the excess antibody present in the immunoinactivated extract that trapped part of the protein (16). In the presence of cordycepin triphosphate, equivalent amounts of the 5′ product were found in wild-type and mutant extracts, indicating that Pab1p is not required for cleavage (data not shown). However, these results show that Pab1p is necessary for the generation of normal-length poly(A) tails in yeast extracts.
What happens with time to the poly(A) tails in the absence or presence of Pab1p? In the pab1Δ rpl46Δ mutant extract alone, the CYC1 precleaved precursor received long poly(A) tails that did not significantly shorten with time (Fig. 5A). These poly(A) tails range from ≈70 to >200 adenylate residues. Within the first 4 min, the precursors received poly(A) tails with an average length of about 110 residues. This is in agreement with previous studies showing that in vivo depletion of Pab1p leads to long poly(A) tails (14). In the mutant extract complemented with Pab1p, the poly(A) tails had an average size of 70 nucleotides already after 1 min (Fig. 5B). However, after 30 min the poly(A) tails became shorter and reached an average size of 40 adenosines after 4 hr. These results indicate that in extracts Pab1p is required for the generation of poly(A) tails of normal length and stimulates a poly(A) shortening activity.
Figure 5.
Kinetics of polyadenylation in the absence or presence of Pab1p. Standard reactions were carried out and stopped at the times indicated. Reaction products were analyzed on 40-cm long 5% polyacrylamide/8 M urea gels. Kinetics of 3′-processing with the pab1Δ rpl46Δ strain (YAS394): (A) in the absence of Pab1p; (B) in the presence of Pab1p (250 ng). M, DNA markers, the sizes are given on the left (in nucleotides). CYC1pre, precleaved precursor; pA, pAn, polyadenylated transcripts.
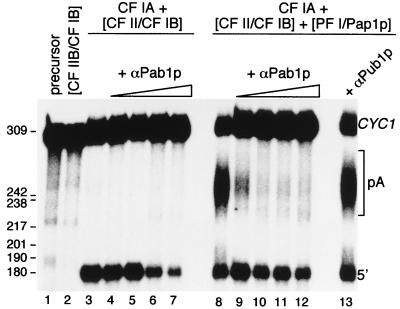
We then wanted to show that Pab1p is also required for polyadenylation in the reconstituted system with purified components. CF IA was preincubated with increasing amounts of the mAb to Pab1p. Low amounts of antibody did not affect cleavage (Fig. 6, lanes 4 and 5) whereas polyadenylation was completely inhibited (Fig. 6, lanes 9 and 10). At a concentration of Pab1p antibody where the polyadenylation reaction is severely inhibited (Fig. 6, lanes 9), a mAb to the poly(U)-binding protein (22) had no effect on the reaction (Fig. 6, lane 13). At higher concentration of anti-Pab1p (Fig. 6, lanes 6, 7, 11, and 12) the cleavage activity also decreased, which is reminiscent of results obtained with extracts (see Fig. 4, lane 3). In this experiment, a complete inactivation of the polyadenylation activity was observed rather than an alteration of the length of the poly(A) tails. The difference between the results obtained with purified factors and with unfractionated pab1Δ mutant extracts could perhaps be explained if one assumes that, at low concentration of antibody, a CF I-Pab1p-antibody complex is made that is still active for cleavage but inapt for polyadenylation. In the absence of Pab1p in the pab1Δ mutant extract, CF I and PF I/Pap1p can still contact each other and polyadenylate the upstream cleavage product but the control of the poly(A) tail length is lost. These data support the conclusion that Pab1p plays an important role in 3′-end processing in yeast.
Figure 6.
Anti-Pab1p inhibition of the reconstituted 3′-end processing reaction with purified factors. Cleavage (with purified CF IA and partially purified CF II/CF IB; lanes 3–7) and cleavage/polyadenylation reactions (with purified CF IA, partially purified CF II/CF IB, and purified PF I/Pap1p; lanes 8–12) were performed with increasing amounts of anti-Pab1p (0, 1.5, 15, 75, and 150 ng, lanes 3–7 and lanes 8–12, respectively). Lane 1, unreacted CYC1 precursor. Lane 2, partially purified CF II/CF IB alone. Lane 13, 1.5 ng of anti-Pub1p is incubated with the cleavage and polyadenylation factors. The sizes of the DNA marker are given on the left (in nucleotides).
DISCUSSION
We report the purification of CF IA from wild-type yeast extracts by complementation of an extract mutant in one of the CF I subunits, Rna15p (10). Although the purification described here differs from that of Kessler et al. (11), it leads to an apparently comparable set of polypeptides of 76, 70, 50, and 40 kDa. We confirmed that the 76- and 40-kDa subunits are Rna14p and Rna15p, respectively (10). We present preliminary data concerning the 50-kDa component that we called Clp1p. Coimmunoprecipitation of Clp1p together with Pab1p, Pcf11p, Rna14p, and Rna15p strongly suggests that we identified the genuine gene coding for the 50-kDa polypeptide (L.M.-S. and W.K., unpublished work).
More remarkably, we showed that the 70-kDa component of CF IA is a mixture of two proteins, Pcf11p (12) and Pab1p (13). Amrani et al. (12) already provided evidence for the involvement of Pcf11p in CF I activity. Our results show that this polypeptide is a component of CF IA. Our data strongly support the idea that in addition to its role in cytoplasmic events (14, 16, 18), Pab1p is also required for the 3′-end formation of mRNAs in the nucleus. Although Pab1p copurifies with CF IA, it is not involved in cleavage, but seems to control the poly(A) tail elongation. Indeed, only poly(A) tails of normal length accumulate in the presence of the poly(A)-binding protein (Fig. 5B), whereas very long polyadenylated species are produced with extracts lacking Pab1p (Fig. 5A).
Formation of the poly(A) tail in mammalian cells also requires the presence of a poly(A)-binding protein, a 49-kDa polypeptide termed PAB II, which is not related to the major 70-kDa poly(A)-binding protein, PAB I, except that they both belong to the RNP-type RNA-binding protein family (for review, see refs. 2 and 25). PAB II has been shown to be involved in the poly(A) tail length control by virtue of a true measurement of the length of the poly(A) tail (26). Earlier studies showed that Pab1p inhibits yeast poly(A) polymerase in vitro (8). The authors suggested that this inhibition could account for the increased poly(A) tail length in pab1 mutants (14). Interestingly, mammalian PAB II has an opposite effect in that it stimulates poly(A) polymerase (27). Because our results showed that Pab1p is essential for 3′-end processing, we can speculate that poly(A) tail length control in yeast is achieved by inhibition of poly(A) polymerase activity. As a working model, we propose that the growing poly(A) tail is progressively bound by Pab1p molecules. Once poly(A) has reached a certain length (≈70 adenosines), the amount of Pab1p bound to it is sufficient to inhibit poly(A) polymerase activity, and subsequent elongation stops. However, the precise mechanism by which the correct poly(A) tail length is adjusted remains to be elucidated. The availability of purified CF I and PF I offers the opportunity to address this question in the future.
As stated before, the fact that apparently only one protein of 70 kDa was seen on the silver-stained gel of the CF IA fraction (Fig. 3, lane 4) would indicate that either Pcf11p or Pab1p has a low affinity for the silver stain. It is also possible that one of the polypeptides is present in undetectable amounts, which would suggest that it is not a tightly associated subunit of CF IA. Interestingly, it has been reported that Pab1p, depending upon the extraction procedure used, is associated with the translation factor eIF-4G (17). This new association of Pab1p with eIF-4F subunits was attributable to the mild conditions of extraction used (17). Similarly, we think for several reasons that Pab1p is not an actual component of CF IA but is rather found loosely associated as an independent factor with the other four subunits. First, the size of the 70 kDa-protein stained with silver fits better that of the band revealed with antibodies to Pcf11p than with anti-Pab1p. Second, Pcf11p behaves more like a bona fide subunit of CF I than Pab1p because, like rna14 and rna15, pcf11 mutants are defective in both cleavage and polyadenylation in vitro (10, 12); on the opposite, pab1 mutant extracts do cleave. Third, in the model we propose, Pab1p is bound to the elongating poly(A) tail. Accordingly, it is not conceivable that Pab1p would stay tightly associated with any factor, but should rather behave like an independent component. With this respect, it is reminiscent of the role of PAB II in the mammalian system (27, 28). For these reasons, we would define Pab1p as a 3′-end processing factor with strong affinity to CF IA. In this view, Pab1p is specifically but transiently associated with CF IA and is being transferred to the growing poly(A) tail during the polyadenylation reaction; it probably also interacts with Pap1p. In any event, our results support the conclusion that Pab1p plays a fundamental role in 3′-end processing in yeast.
We have also shown that only in the presence of Pab1p the poly(A) tails are shortened (Fig. 5B). This result is in apparent contradiction to the finding that poly(A) shortening in vivo does not require Pab1p, although in its absence deadenylation rates are reduced (18). However, one can reconcile this discrepancy by assuming that our in vitro data reproduce a requirement for Pab1p in an initial shortening that already takes place in the nucleus. This reaction is probably different from the cytoplasmic deadenylation that precedes mRNA degradation (29, 30). It would be interesting to test whether the recently identified Pab1p-dependent poly(A) nuclease (19, 20) participates in the control of poly(A) tail length in the nucleus. In this model, Pab1p would have a two-fold effect in that it would inhibit poly(A) polymerase activity and activate the poly(A) nuclease.
Acknowledgments
We thank Alan Sachs for materials and comments, Maurice Swanson and François Lacroute for antibodies, and Katrin Beyer, Mary O’Connell, and Ursula Rüegsegger for critically reading the manuscript and suggestions. We are especially indebted to Elmar Wahle and Alan Sachs for many fruitful discussions and comments. This work was supported by grants from the Kantons of Basel, the Swiss National Science Foundation, and the European Union via the Bundesamt für Bildung und Wissenschaft (Bern).
ABBREVIATIONS
- CF
cleavage factor
- PF
polyadenylation factor
- Pap1p
poly(A) polymerase
- Pab1p
poly(A)-binding protein
- Clp1p
cleavage/polyadenylation factor IA subunit
- Pcf11p
protein 1 of CF I. PAB I and II, 70- and 49-kDa poly(A)-binding protein, respectively
References
- 1.Wahle E, Keller W. Trends Biochem Sci. 1996;21:247–250. [PubMed] [Google Scholar]
- 2.Wahle E, Keller W. Annu Rev Biochem. 1992;61:419–440. doi: 10.1146/annurev.bi.61.070192.002223. [DOI] [PubMed] [Google Scholar]
- 3.Manley J L. Curr Opin Genet Dev. 1995;5:222–228. doi: 10.1016/0959-437x(95)80012-3. [DOI] [PubMed] [Google Scholar]
- 4.Guo Z, Sherman F. Trends Biochem Sci. 1996;21:477–481. doi: 10.1016/s0968-0004(96)10057-8. [DOI] [PubMed] [Google Scholar]
- 5.Butler J S, Platt T. Science. 1988;242:1270–1274. doi: 10.1126/science.2848317. [DOI] [PubMed] [Google Scholar]
- 6.Butler J S, Sadhale P P, Platt T. Mol Cell Biol. 1990;10:2599–2605. doi: 10.1128/mcb.10.6.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J, Moore C. Mol Cell Biol. 1992;12:3470–3481. doi: 10.1128/mcb.12.8.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lingner J, Radkte I, Wahle E, Keller W. J Biol Chem. 1991;266:8741–8746. [PubMed] [Google Scholar]
- 9.Lingner J, Kellermann J, Keller W. Nature (London) 1991;354:496–498. doi: 10.1038/354496a0. [DOI] [PubMed] [Google Scholar]
- 10.Minvielle-Sebastia L, Preker P J, Keller W. Science. 1994;266:1702–1705. doi: 10.1126/science.7992054. [DOI] [PubMed] [Google Scholar]
- 11.Kessler M M, Zhao J, Moore C L. J Biol Chem. 1996;271:27167–27175. doi: 10.1074/jbc.271.43.27167. [DOI] [PubMed] [Google Scholar]
- 12.Amrani N, Minet M, Wyers F, Dufour M-E, Aggerbeck L P, Lacroute F. Mol Cell Biol. 1997;17:1102–1109. doi: 10.1128/mcb.17.3.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sachs A B, Bond M W, Kornberg R D. Cell. 1986;45:827–835. doi: 10.1016/0092-8674(86)90557-x. [DOI] [PubMed] [Google Scholar]
- 14.Sachs A B, Davis R W. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 15.Sachs A, Deardorff J A. Cell. 1992;70:961–973. doi: 10.1016/0092-8674(92)90246-9. [DOI] [PubMed] [Google Scholar]
- 16.Tarun S J, Sachs A B. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 17.Tarun S Z, Sachs A B. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 18.Caponigro G, Parker R. Genes Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- 19.Boeck R, Tarun S, Rieger M, Deardorff J A, Müller-Auer S, Sachs A B. J Biol Chem. 1996;271:432–438. doi: 10.1074/jbc.271.1.432. [DOI] [PubMed] [Google Scholar]
- 20.Brown C E, Tarun S Z, Boeck R, Sachs A B. Mol Cell Biol. 1996;16:5744–5753. doi: 10.1128/mcb.16.10.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sachs A B, Davis R W, Kornberg R D. Mol Cell Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson J T, Paddy M R, Swanson M S. Mol Cell Biol. 1993;13:6102–6113. doi: 10.1128/mcb.13.10.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Preker P J, Lingner J, Minvielle-Sebastia L, Keller W. Cell. 1995;81:379–389. doi: 10.1016/0092-8674(95)90391-7. [DOI] [PubMed] [Google Scholar]
- 24.Preker, P. J., Ohnacker, M., Minvielle-Sebastia, L. & Keller, W. (1997) EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 25.Keller W, Minvielle-Sebastia L. Curr Opin Cell Biol. 1997;9:329–336. doi: 10.1016/s0955-0674(97)80004-x. [DOI] [PubMed] [Google Scholar]
- 26.Wahle E. J Biol Chem. 1995;270:2800–2808. doi: 10.1074/jbc.270.6.2800. [DOI] [PubMed] [Google Scholar]
- 27.Wahle E. Cell. 1991;66:759–768. doi: 10.1016/0092-8674(91)90119-j. [DOI] [PubMed] [Google Scholar]
- 28.Wahle E, Lustig A, Jenö P, Maurer P. J Biol Chem. 1993;268:2937–2945. [PubMed] [Google Scholar]
- 29.Decker C J, Parker R. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 30.Decker C J, Parker R. Trends Biochem Sci. 1994;19:336–340. doi: 10.1016/0968-0004(94)90073-6. [DOI] [PubMed] [Google Scholar]