Abstract
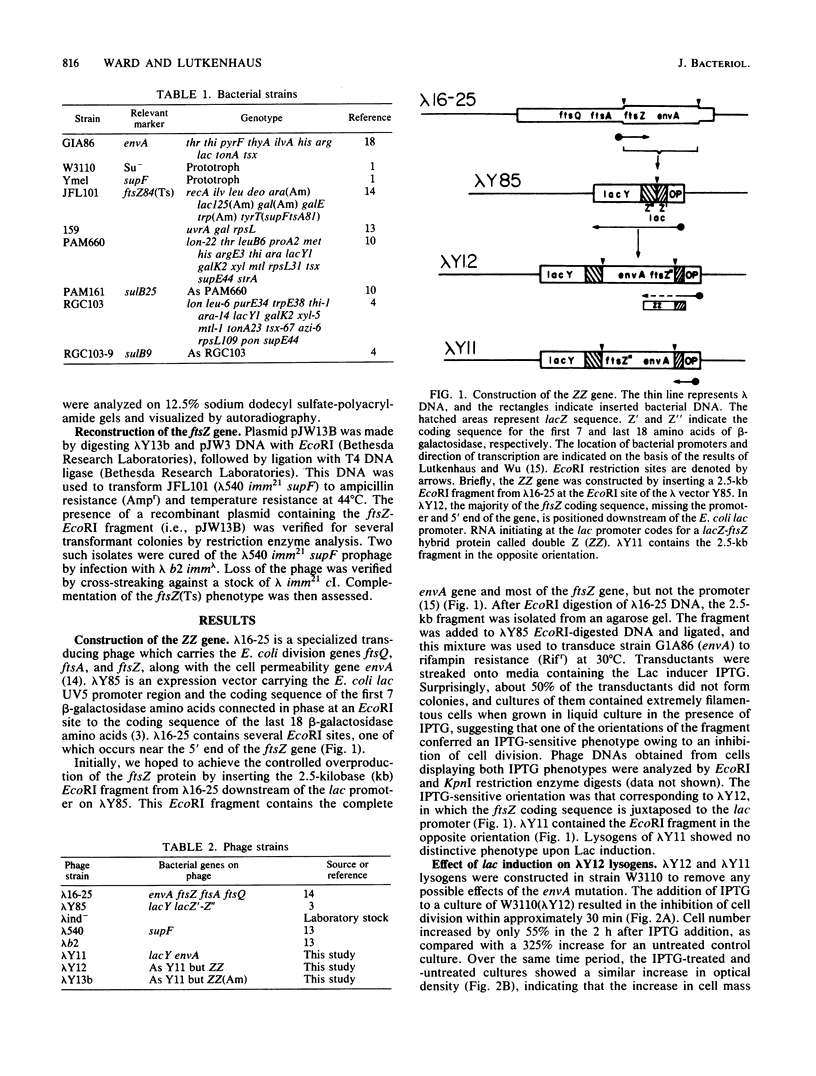
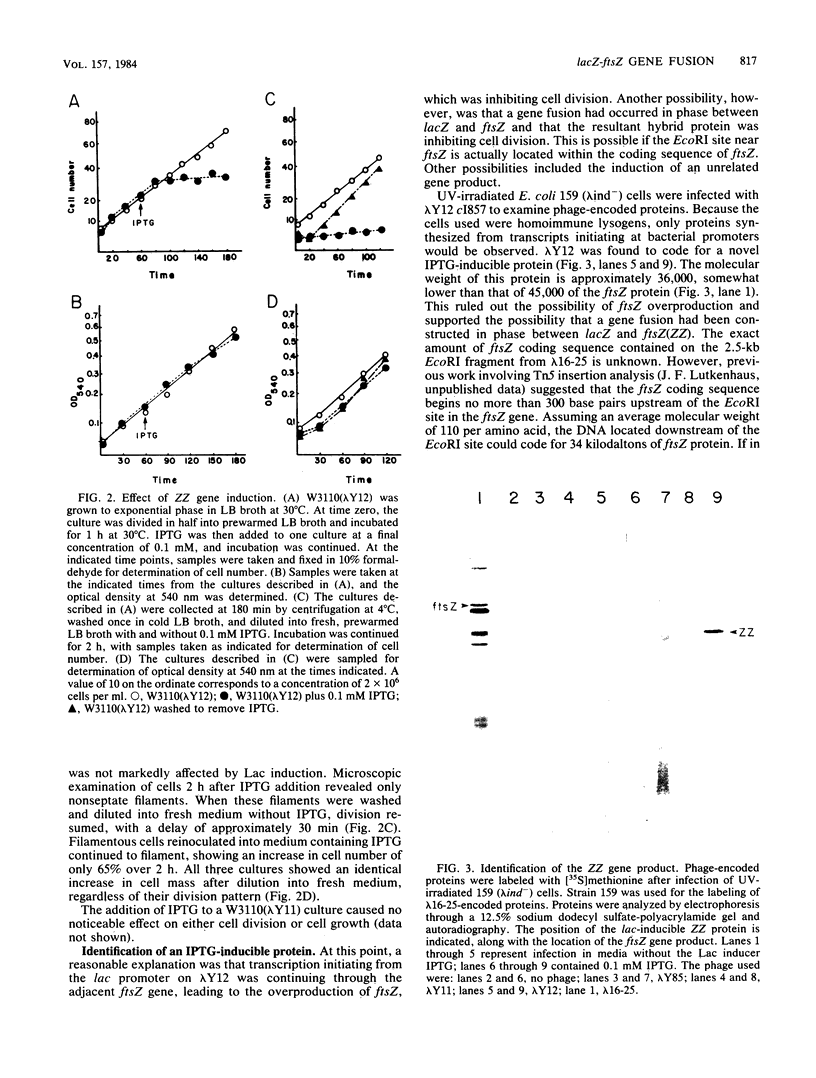
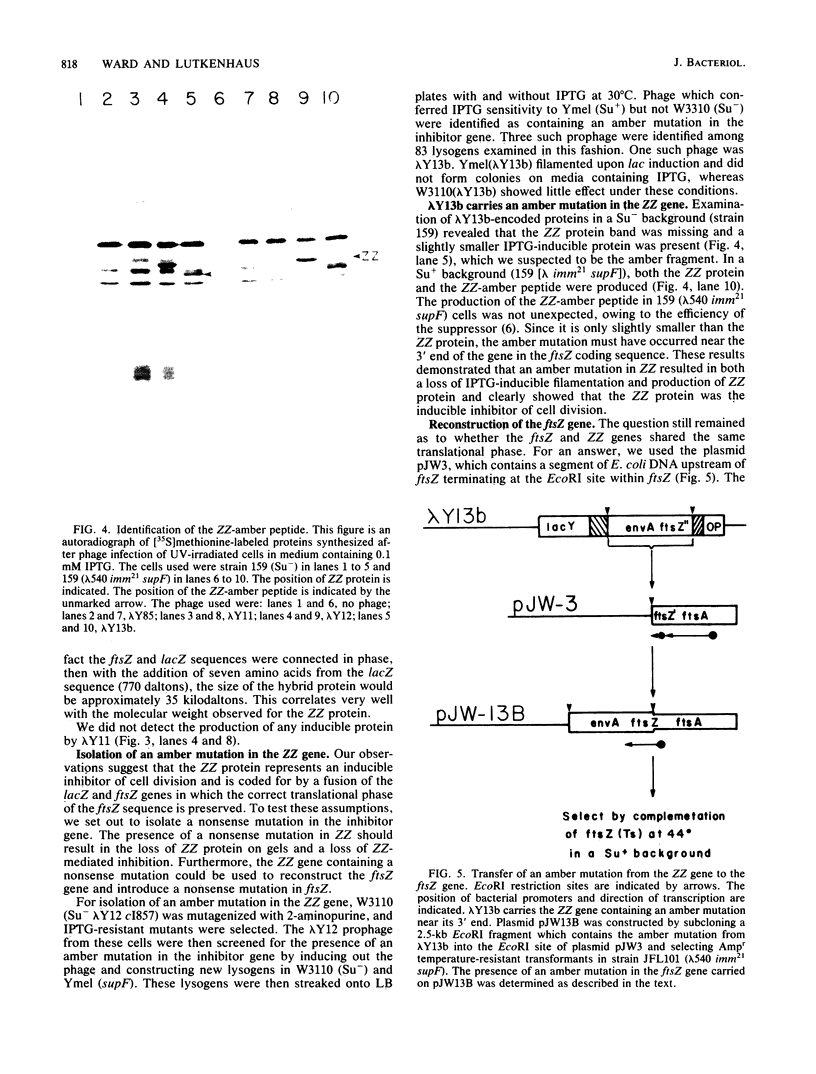
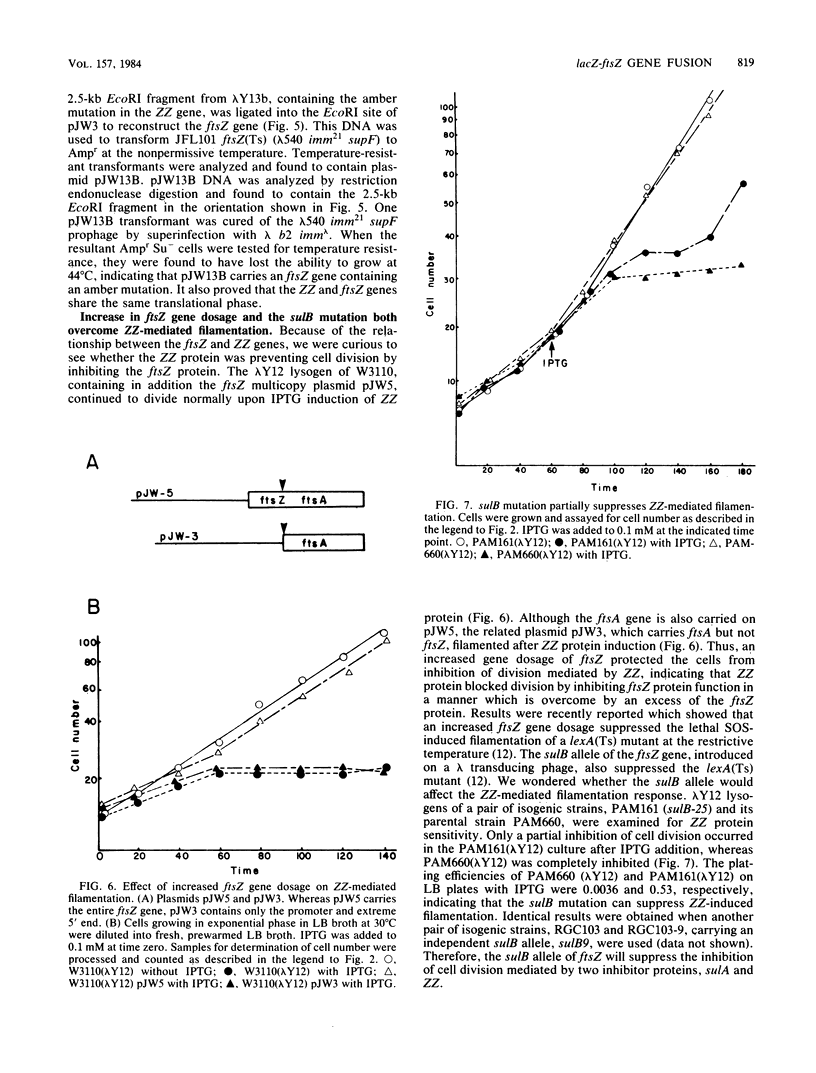
An in-frame lacZ-ftsZ gene fusion under lac control was fortuitously constructed by subcloning an EcoRI fragment that contains approximately 90% of the ftsZ gene. The identity of the gene fusion was confirmed by isolating an amber mutation in the hybrid gene and then using it to reconstruct the ftsZ gene, which now contained an amber mutation. The hybrid protein (ZZ), which does not possess ftsZ activity, contains seven amino acids of lacZ at its amino terminal end, followed by 35,000 daltons of the carboxyl end of the ftsZ protein. Induction of the hybrid protein resulted in a rapid cessation of cell division which could be reversed by removing the lac inducer. This inhibition of division could be prevented by an increased gene dosage of ftsZ or the presence of the sulB allele of ftsZ, which is known to code for an altered but functional ftsZ protein. An increased gene dosage of ftsZ or the presence of the sulB allele of ftsZ is known to overcome sulA-mediated inhibition of division during the SOS response. Thus, our results suggest that ZZ is an analog of sulA and may aid in determining how sulA inhibits cell division.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Borck K., Beggs J. D., Brammar W. J., Hopkins A. S., Murray N. E. The construction in vitro of transducing derivatives of phage lambda. Mol Gen Genet. 1976 Jul 23;146(2):199–207. doi: 10.1007/BF00268089. [DOI] [PubMed] [Google Scholar]
- Burdett I. D., Murray R. G. Septum formation in Escherichia coli: characterization of septal structure and the effects of antibiotics on cell division. J Bacteriol. 1974 Jul;119(1):303–324. doi: 10.1128/jb.119.1.303-324.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charnay P., Louise A., Fritsch A., Perrin D., Tiollais P. Bacteriophage lambda-E. coli K12 vector-host system for gene cloning and expression under lactose promoter control. II. DNA fragment insertion at the vicinity of the lac UV5 promoter. Mol Gen Genet. 1979 Feb 26;170(2):171–178. doi: 10.1007/BF00337793. [DOI] [PubMed] [Google Scholar]
- Gayda R. C., Yamamoto L. T., Markovitz A. Second-site mutations in capR (lon) strains of Escherichia coli K-12 that prevent radiation sensitivity and allow bacteriophage lambda to lysogenize. J Bacteriol. 1976 Sep;127(3):1208–1216. doi: 10.1128/jb.127.3.1208-1216.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J., Castellazzi M., Buttin G. Prophage induction and cell division in E. coli. III. Mutations sfiA and sfiB restore division in tif and lon strains and permit the expression of mutator properties of tif. Mol Gen Genet. 1975 Oct 22;140(4):309–332. [PubMed] [Google Scholar]
- Gorini L. Informational suppression. Annu Rev Genet. 1970;4:107–134. doi: 10.1146/annurev.ge.04.120170.000543. [DOI] [PubMed] [Google Scholar]
- Gottesman S., Halpern E., Trisler P. Role of sulA and sulB in filamentation by lon mutants of Escherichia coli K-12. J Bacteriol. 1981 Oct;148(1):265–273. doi: 10.1128/jb.148.1.265-273.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman O., D'Ari R., George J. Further characterization of sfiA and sfiB mutations in Escherichia coli. J Bacteriol. 1980 Oct;144(1):185–191. doi: 10.1128/jb.144.1.185-191.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman O., D'Ari R., George J. Inducible sfi dependent division inhibition in Escherichia coli. Mol Gen Genet. 1980;177(4):629–636. doi: 10.1007/BF00272673. [DOI] [PubMed] [Google Scholar]
- Johnson B. F. Fine structure mapping and properties of mutations suppressing the lon mutation in Escherichia coli K-12 and B strains. Genet Res. 1977 Dec;30(3):273–286. doi: 10.1017/s0016672300017687. [DOI] [PubMed] [Google Scholar]
- Little J. W., Mount D. W. The SOS regulatory system of Escherichia coli. Cell. 1982 May;29(1):11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. F. Coupling of DNA replication and cell division: sulB is an allele of ftsZ. J Bacteriol. 1983 Jun;154(3):1339–1346. doi: 10.1128/jb.154.3.1339-1346.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. F., Donachie W. D. Identification of the ftsA gene product. J Bacteriol. 1979 Mar;137(3):1088–1094. doi: 10.1128/jb.137.3.1088-1094.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. F., Wolf-Watz H., Donachie W. D. Organization of genes in the ftsA-envA region of the Escherichia coli genetic map and identification of a new fts locus (ftsZ). J Bacteriol. 1980 May;142(2):615–620. doi: 10.1128/jb.142.2.615-620.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. F., Wu H. C. Determination of transcriptional units and gene products from the ftsA region of Escherichia coli. J Bacteriol. 1980 Sep;143(3):1281–1288. doi: 10.1128/jb.143.3.1281-1288.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. R., Kovarik A., Allen J. S., Gustafson R. A. Regulation of bacterial cell division: temperature-sensitive mutants of Escherichia coli that are defective in septum formation. J Bacteriol. 1975 Aug;123(2):693–703. doi: 10.1128/jb.123.2.693-703.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijsman H. J., Koopman C. R. The relation of the genes envA and ftsA in Escherichia coli. Mol Gen Genet. 1976 Aug 10;147(1):99–102. doi: 10.1007/BF00337942. [DOI] [PubMed] [Google Scholar]
- Witkin E. M. Ultraviolet mutagenesis and inducible DNA repair in Escherichia coli. Bacteriol Rev. 1976 Dec;40(4):869–907. doi: 10.1128/br.40.4.869-907.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]





