Abstract
The restriction factors, TRIM5α in most primates and TRIMCyp in owl monkeys, block infection of various retroviruses soon after virus entry into the host cell. Rhesus monkey TRIM5α (TRIM5αrh) inhibits human immunodeficiency virus (HIV-1) and feline immunodeficiency virus (FIV) more potently than human TRIM5α (TRIM5αhu). TRIMCyp restricts infection of HIV-1, simian immunodeficiency virus of African green monkeys (SIVagm) and FIV. Early after infection, TRIMCyp, like TRIM5αrh and TRIM5αhu, decreased the amount of particulate viral capsid in the cytosol of infected cells. The requirements for the TRIMCyp and TRIM5α domains in restricting different retroviruses were investigated. Potent restriction of FIV by TRIMCyp occurred in the complete absence of RING and B-box 2 domains; by contrast, efficient FIV restriction by TRIM5αrh required these domains. Variable region 1 of the TRIM5αrh B30.2 domain contributed to the potency of HIV-1, FIV and equine infectious anemia virus restriction. Thus, although differences exist in the requirements of TRIMCyp and TRIM5α for RING/B-box 2 domains, both restriction factors exhibit mechanistic similarities.
Keywords: Restriction factors, retrovirus, uncoating, human immunodeficiency virus
Introduction
Following entry into cells, some retroviruses encounter dominant blocks to infection that act prior to reverse transcription (Cowan et al., 2002; Munk et al., 2002). For example, human immunodeficiency virus (HIV-1) is blocked in the cells of most Old World monkeys (Hofmann et al., 1999). Infection by the simian immunodeficiency virus of macaques (SIVmac) encounters similar restrictions in New World monkeys (Hofmann et al., 1999). These restrictions are mediated by TRIM5α, a tripartite motif protein with a RING, B-box 2 and coiled-coil (CC) domain, as well as a carboxy-terminal B30.2(SPRY) domain (Stremlau et al., 2004). The TRIM5α B30.2 domain is essential for retroviral restriction, and differences in the potency of TRIM5α from different species map to this domain (Sawyer et al., 2005; Stremlau et al., 2005; Yap, Nisole, and Stoye, 2005). In addition to inhibiting HIV-1, TRIM5αrh potently restricts feline immunodeficiency virus (FIV) and equine infectious anemia virus (EIAV) infections (Hatziioannou et al., 2004; Keckesova, Ylinen, and Towers, 2004; Olsen, 1998; Saenz et al., 2005).
Owl monkeys are an unusual New World monkey species that exhibit restrictions to HIV-1 and not SIVmac infection (Hofmann et al., 1999; Towers et al., 2003). Instead of TRIM5α, owl monkeys express TRIMCyp, a protein that consists of the RING, B-box 2 and CC domains of TRIM5 fused with a carboxy-terminal cyclophilin A (Cyp A) moiety (Nisole et al., 2004; Sayah et al., 2004; Ribeiro et al., 2005). The TRIMCyp gene was generated by a retrotransposition event involving the TRIM5 and Cyp A genes. Cyp A is known to bind a surface-exposed loop on the HIV-1 p24 capsid protein (Gamble et al., 1996; Luban et al., 1993). Besides restricting HIV-1 infection, TRIMCyp also potently inhibits infection by FIV and SIVagm (Diaz-Griffero et al., 2006b; Lin and Emerman, 2006).
The viral determinant of susceptibility to TRIM5α- and TRIMCyp-mediated restriction is the capsid protein, which assembles into the capsid structure that surrounds the viral genomic RNA and enzymes (Owens et al., 2004; Owens et al., 2003). Studies following the fate of the HIV-1 and N-MLV capsids in the cytoplasm of cells expressing TRIM5αrh or TRIM5αhu, respectively, established a correlation between restriction and the premature conversion of particulate capsids to soluble capsid proteins (Perron et al., 2007; Stremlau et al., 2006). However, the effect of TRIMCyp on the fate of restricted retroviral capsids is not known.
Here we investigate the requirements for retroviral restriction by TRIM5αrh and TRIMCyp. Like TRIM5αrh, TRIMCyp decreased the amount of particulate capsid in the cytosol following HIV-1 infection. As TRIMCyp and TRIM5αrh potently restrict FIV infection, we determined the contribution of each TRIM domain to this restriction. FIV restriction by TRIMCyp did not require the RING and B-box 2 domains for full potency. In contrast, TRIM5αrh required the RING and B-box 2 sequences to achieve efficient restriction of FIV. Studies using human-monkey TRIM5α chimerae demonstrated that the variable region 1 of the B30.2 domain contains potency determinants for FIV restriction.
Results
Effect of TRIMCyp on the amount of particulate capsid during HIV-1 infection
Both TRIM5αrh and TRIMCyp potently restrict HIV-1 infection. The expression of TRIM5αrh in target cells results in a decrease in the amount of particulate cytosolic capsids during HIV-1 infection (Stremlau et al., 2006). To test the effect of TRIMCyp on the integrity of HIV-1 capsid complexes in the cytosol of infected cells, we studied the fate of the HIV-1 capsid in Cf2Th canine thymocytes expressing TRIMCyp and TRIM5αrh. As shown in Figure 1A, expression of TRIMCyp reduced the amount of cytosolic particulate capsid compared with that in cells transduced with the empty LPCX vector. The decrease in the amount of particulate HIV-1 capsid in the cytosol of cells expressing TRIMCyp was comparable to that observed as a result of TRIM5αrh expression. No cytosolic capsid was detected in cells challenged with HIV-1 particles lacking envelope glycoproteins. Similar results were obtained after HIV-1 challenge of CRFK feline cells expressing TRIMCyp and TRIM5αrh (Figure 1B). Deletion of the TRIMCyp RING and B-box 2 domains (TRIMCypΔ128) or alteration of the TRIMCyp B- box 2 domain (TRIMCyp R120E) abrogated the TRIMCyp-mediated decrease in the level of cytosolic, particulate capsids (Figure 1C, upper panel). These observations suggest that the expression of TRIMCyp, like that of TRIM5αrh (Stremlau et al., 2006), results in a decrease in the amount of particulate capsid in the cytosol of HIV-1-infected cells.
Figure 1. Fate of the HIV-1 capsid in the cytosol of cells expressing TRIM5αrh and TRIMCyp.
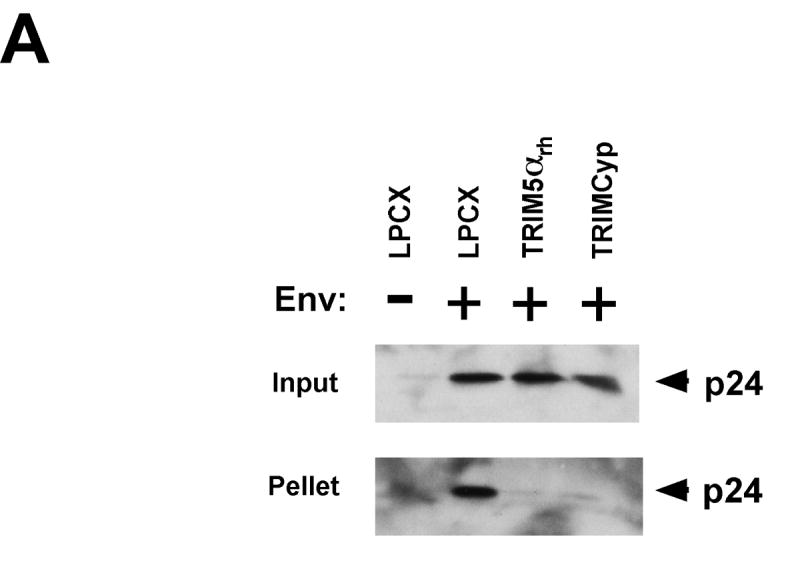
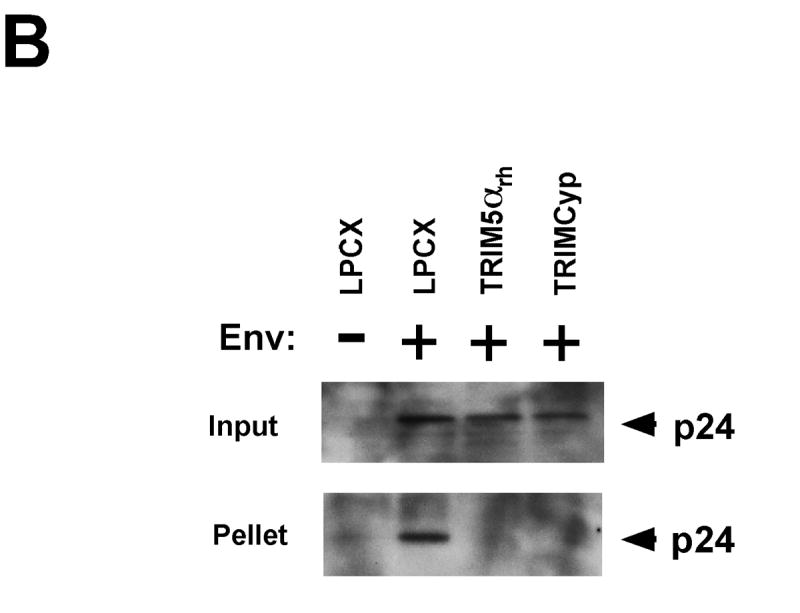



Cf2Th (A, C, D) or CRFK (B) cells transduced with TRIMCyp, TRIMCyp Δ128, TRIMCyp R120E, TRIM5αrh or the empty LPCX control vector were incubated with either VSV G-pseudotyped (Env+) HIV-1 or envelope-deficient (Env-) HIV-1 for 16 hours in the presence or absence of 10 μM MG115. Cytoplasmic lysates were analyzed directly (“Input”) or were separated on a sucrose gradient into soluble and particulate fractions. The particulate fractions (“Pellet”) were analyzed by Western blot using an antibody directed against the HIV-1 p24 capsid protein. In E, Cf2Th cells expressing TRIM5αrh and TRIMCyp were treated for 16 hours with either DMSO or 10 μM MG115. The cells were then fixed and stained with an antibody directed against the HA epitope tag.
Proteasome inhibition does not affect the efficiency of TRIM5αrh–mediated restriction of HIV-1 (Perez-Caballero et al., 2005b; Stremlau et al., 2006), but has been reported to rescue the production of viral reverse transcripts in cells expressing a restricting TRIM5α protein (Anderson et al., 2006; Wu et al., 2006). We examined the effects of proteasome inhibition on the TRIM5αrh– and TRIMCyp–mediated decrease in the level of particulate, cytosolic HIV-1 capsids in infected cells. Target cells expressing TRIM5αrh or TRIMCyp, or control cells transduced with the empty LPCX vector, were treated with the proteasome inhibitor MG115 throughout the period of exposure to HIV-1. In the control cells as well as the cells expressing TRIM5αrh or TRIMCyp, a dramatic increase in the amount of pelletable cytosolic HIV-1 capsid was observed after MG115 treatment (Figure 1, C and D). No significant differences were observed between theamounts of cytosolic, pelletable capsid in the control or TRIM5αrh/TRIMCyp-expressing cells in the presence of MG115. Thus, proteasome inhibition results in a general increase in the amount of particulate capsids in the cytosol of HIV-1-infected cells.
To examine the effect of proteasome inhibition on the intracellular distribution of the TRIM5αrh and TRIMCyp proteins, cells stably expressing these proteins were treated with MG115 or the DMSO solvent. The cells were then stained with an antibody directed against the hemagglutinin (HA) epitope tag on the TRIM5αrh and TRIMCyp proteins. In the cells treated with the DMSO solvent, TRIM5αrh and TRIMCyp exhibited diffuse cytoplasmic staining punctuated by cytoplasmic speckles (Figure 1E). By contrast, the TRIM5αrh and TRIMCyp proteins in the MG115-treated cells coalesced into large inclusion bodies. These results are consistent with previous observations suggesting that proteasome inhibition leads to the movement of TRIM5α into pre- aggresomal cytoplasmic bodies (Diaz-Griffero et al., 2006a).
Timing of the TRIMCyp effect on particulate cytosolic HIV-1 capsids
The owl monkey TRIMCyp protein acts to restrict HIV-1 infection within the first hour following virus entry (Diaz-Griffero et al., 2006b; Perez-Caballero et al., 2005b). To examine the kinetics of the TRIMCyp-mediated decrease in cytosolic levels of particulate capsid, we challenged control cells or TRIMCyp-expressing cells with HIV-1 pseudotyped with the VSV-G envelope glycoprotein and followed the amount of particulate capsid over a 48-hour period. At all time points examined, the amount of particulate capsid in TRIMCyp-expressing cells was less than that seen in the LPCX-transduced control cells (Figure 2). No capsid protein was observed in cell lysates prepared 16 hours after challenge of TRIMCyp-expressing or LPCX-transduced cells with HIV-1 lacking envelope glycoproteins. Thus, the amount of pelletable capsid in the cytosol of LPCX-transduced cells is greater than that in TRIMCyp-expressing cells at multiple time points after HIV-1 infection.
Figure 2. Time course of TRIMCyp-induced loss of particulate HIV-1 capsids in infected cells.

Cf2Th cells transduced with either an empty LPCX vector or an LPCX vector expressing TRIMCyp were incubated at 4°C for 30 minutes with either HIV-1 Env+ or Env- virus-like particles. Cells were then incubated at 37°C for the indicated times and lysed. Cytoplasmic lysates were analyzed directly (“Input”) or were separated on a sucrose gradient into soluble and particulate fractions. The particulate fractions (“Pellet”) were Western blotted using an antibody directed against the HIV-1 p24 protein.
TRIMCyp domains involved in the restriction of feline immunodeficiency virus (FIV) infection
Owl monkey TRIMCyp blocks FIV and SIVagm infection in addition to that of HIV-1 (Diaz-Griffero et al., 2006b; Lin and Emerman, 2006). To examine the role of the different TRIMCyp domains in the restriction of FIV infection, we stably expressed a previously described set of TRIMCyp variants with different N-terminal deletions in CRFK feline cells (Figures 3A and 3B). Deletion of the RING domain or RING and B-Box 2 domains of TRIMCyp (TRIMCypΔ93 and TRIMCypΔ128, respectively) did not affect the ability of the protein to restrict FIV infection efficiently (Figure 3C). In contrast, similar deletions partially decreased the ability of the TRIMCyp protein to restrict HIV-1 and SIVagm infection (Figures 3D and 3E).
Figure 3. FIV restriction by TRIMCyp variants.



CRFK cells were transduced with either the empty LPCX vector or an LPCX vector expressing wild-type TRIMCyp or the TRIMCyp mutants shown in A. Cell lysates were analyzed by Western blot using antibodies against HA (upper panel) and β-actin (lower panel) (B). CRFK cells expressing the different TRIMCyp variants were challenged with FIV-GFP (C), HIV-1-GFP (D) or SIVagm-GFP (E). GFP-positive cells were counted by flow cytometry. Similar results were obtained in three independent experiments.
Deletion of the TRIMCyp coiled coil in the TRIMCypΔ245 and TRIMCypΔ310 mutants resulted in the complete elimination of FIV-restricting ability (Figure 3C). These mutants exhibited weak inhibitory activity against HIV-1 and SIVagm (Figures 3D and 3E). Thus, as has been previously observed (Diaz-Griffero et al., 2006b; Hatziioannou et al., 2005; Yin, Braaten, and Luban, 1998), overexpression of cyclophilin A-like proteins can result in decreased susceptibility of cells to HIV-1.
TRIM5αrh domains involved in the restriction of FIV infection
FIV infection is also potently blocked by TRIM5αrh (Saenz et al., 2005). To examine if FIV restriction requires the TRIM5αrh RING and B-box 2 domains, we expressed wild-type TRIM5αrh and TRIM5αrh mutants with N-terminal deletions in CRFK feline cells (Figures 4A and 4B). The cells were challenged with FIV (Figure 4C). In contrast to the results obtained for TRIMCyp, deletions of the RING and B-box 2 domains dramatically attenuated the ability of TRIM5αrh to inhibit FIV infection.
Figure 4. FIV restriction by TRIM5αrh.

CRFK cells expressing the indicated wild-type and mutant TRIM5αrh proteins (A, B) were challenged with FIV-GFP (C). GFP-positive cells were counted. Similar results were obtained in three independent experiments.
TRIM5αrh potency against FIV maps to the V1 region of the B30.2 domain
The TRIM5α B30.2 domain determines the specificity of recognition of the targeted retroviral capsid (Saenz et al., 2005; Song et al., 2005a; Stremlau et al., 2005). The v1 variable region of the B30.2 domain of TRIM5αrh is the major determinant of anti-HIV-1 potency (Li et al., 2006; Perez-Caballero et al., 2005a; Stremlau et al., 2005; Yap, Nisole, and Stoye, 2005). Sequences in the v1 and v3 regions of the TRIM5αhu B30.2 domain contribute to efficient N-MLV restriction (Perron, Stremlau, and Sodroski, 2006). To investigate the TRIM5α determinants of potency for FIV restriction, we took advantage of the observation that TRIM5αrh efficiently restricts FIV infection, whereas TRIM5αhu only weakly inhibits FIV infection. Protein chimerae containing different segments of TRIM5αrh and TRIM5αhu (Stremlau et al., 2005) were expressed in CRFK cells (Figures 5A and 5B). The chimeric proteins all formed trimers (See Figure 7 below). A chimeric protein (H(R286-371)), in which TRIM5αhu amino acids 286-371 have been replaced by the equivalent sequences of TRIM5αrh, inhibited FIV infection comparably to TRIM5αrh (Figure 5C). The reciprocal construct, R(H286-371), in which a B30.2(SPRY) domain segment spanning the v1 region of TRIM5αrh has been replaced by that of TRIM5αhu, exhibited no inhibition of FIV infection. These results suggest that the B30.2(SPRY) domain segment that includes the v1 variable region determines the potency of FIV restriction.
Figure 5. TRIM5αrh potency against FIV maps to the v1 region of the B30.2 domain.

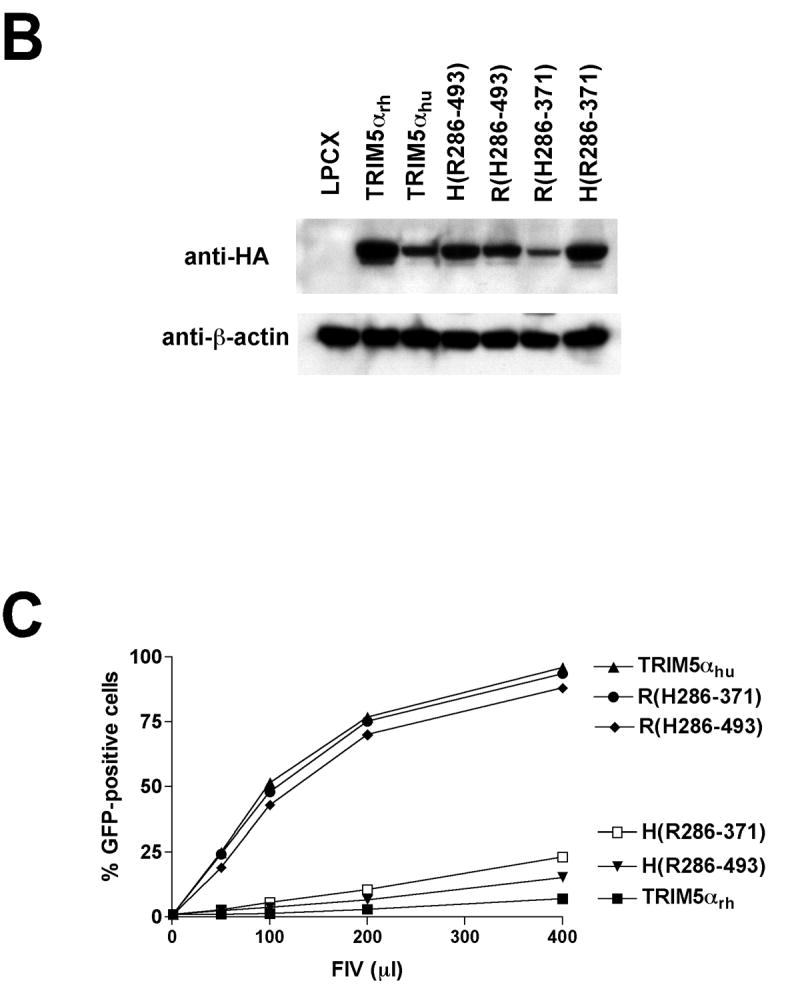
CRFK cells expressing the indicated wild-type and TRIM5αrh-TRIM5αhu chimeric proteins (A) were Western blotted with an antibody directed against the HA epitope tag and control anti-β-actin antibody (B). Cells were challenged with FIV-GFP (C). GFP-positive cells were counted. Similar results were obtained in three independent experiments.
Figure 7. Oligomerization of TRIM5α variants.

293T cells were transiently transfected with plasmids expressing TRIM5α variants. Cells were lysed 36 hours after transfection and crosslinked with the indicated concentrations of ethylene glycol-bis(succinimidyl succinate) (EGS) as described in Materials and Methods. The cell lysates were subsequently blotted with an anti-HA antibody.
Effects of changes in the TRIM5α B30.2 domain v1 region on retrovirus restriction
The effects of deletions in the B30.2 domain v1 region on retroviral restriction by TRIM5αrh were examined. TRIM5αrh mutants with deletions affecting different segments of the B30.2 v1 region (Figure 6A) were stably expressed in CRFK cells (Figure 6B). The cells were challenged with recombinant HIV-1, EIAV and FIV expressing GFP (Figure 6C, D and E). A short deletion (Δ333-339) in the middle of the v1 variable region exerted only minimal effects on the ability of TRIM5αrh to restrict these three retroviruses, even though the expression level of the mutant TRIM5αrh protein was relatively low. A larger deletion (Δ332-344) in the B30.2 v1 region significantly decreased TRIM5αrh restricting activity. Intermediate levels of restriction were observed for proteins [TRIM5αrh (Δ323-332) and TRIM5αrh (Δ332-344)] with deletions affecting the flanks of the B30.2 v1 region. A deletion of the B30.2 v3 region (Δ403-440) eliminated detectable restricting activity of TRIM5αrh without affecting the oligomerization state of the protein (Figure 7). These results support a role of the B30.2 variable regions in the restriction of HIV-1, EIAV and FIV infection.
Figure 6. Effects of changes in the TRIM5αrh and TRIM5αhu B30.2 domain v1 region on restriction of HIV-1, EIAV and FIV.





(A) Wild-type TRIM5αrh and TRIM5αhu and the indicated mutants were expressed in CRFK cells. Cells were lysed and Western blotted with an anti-HA antibody or a control antibody directed against β-actin (B). Cells were challenged with recombinant HIV-1 (C), EIAV (D) or FIV (E, G) expressing GFP. GFP-positive cells were counted. Similar results were obtained in a repeat experiment. (F) Cell lysates containing the indicated TRIM5αrh variants were incubated with HIV-1 CA-NC complexes. The mixtures were layered on a 70% sucrose cushion and centrifuged. The input lysates and pellets were Western blotted with an antibody directed against the TRIM5αrh HA epitope tag. The pellets were also Western blotted with an anti-p24 CA antibody.
To investigate the mechanism by which B30.2 v1 region deletions affect restricting activity, the ability of the TRIM5αrh mutants to bind HIV-1 capsid-nucleocapsid (CA-NC) complexes was examined (Figure 6F). The wild-type TRIM5αrh protein and the potent restriction factor TRIM5αrh Δ(333-339) efficiently associated with HIV-1 CA-NC complexes. The other TRIM5αrh mutants tested bound much less efficiently to the HIV-1 CA-NC preparations. Thus, decreased ability to bind the HIV-1 capsids contributes to the diminished HIV-1-restricting activity of TRIM5αrh mutants with deletions affecting the B30.2(SPRY) v1 region.
Changes in a single residue, arginine 332, within the B30.2 v1 region of TRIM5αhu can create a potent restrictor of HIV-1 infection (Stremlau et al., 2005; Yap, Nisole, and Stoye, 2005; Li et al., 2006). To examine whether this residue affects the ability of TRIM5αhu to inhibit FIV infection, cells expressing TRIM5αhu R332P were challenged with FIV-GFP. Figure 6G shows that TRIM5αhu R332P inhibited FIV infection comparably to the wild-type TRIM5αhu protein. The H(R323-332) protein, a TRIM5αhu variant that contains residues 323-332 from TRIM5αhu (Figure 6A), restricted FIV infection more efficiently than the wild-type TRIM5αhu protein. Thus, the determinants of TRIM5α potency for the restriction of HIV-1 and FIV infection are closely related but not identical, and include the v1 region of the B30.2(SPRY) domain.
Oligomerization of TRIM5α variants
TRIM5α assembles into trimers, a property that contributes to its ability to bind HIV-1 capsids and to restrict retroviral infection (Mische et al., 2005; Javanbakht et al., 2006). To examine the oligomerization of the TRIM5α variants studied herein, lysates from cells expressing these proteins were crosslinked and analyzed (Figure 7). All of the TRIM5α proteins examined formed trimers.
Discussion
The steady-state level of capsid protein in the cytosol of HIV-1-infected cells is determined by the amount of capsid entering the cell and the amount of capsid protein undergoing degradation. The total levels of HIV-1 capsid protein were similar in the cytosol of infected control cells and cells expressing TRIMCyp or TRIM5αrh. This observation is consistent with a minimal effect of TRIMCyp or TRIM5αrh on HIV-1 entry or capsid turnover.
Compared with those in control cells, the levels of particulate cytosolic HIV-1 capsids in TRIMCyp- or TRIM5αrh-expressing cells were decreased. The results observed herein are consistent with previously described models in which restriction factors promote the premature uncoating of the incoming retroviral capsid (Perron et al., 2007; Stremlau et al., 2006). Despite differences in the capsid-binding moieties of TRIMCyp and TRIM5α, the ultimate consequences of binding these factors to the HIV-1 capsid appear to be remarkably similar.
Differences exist in the requirements for the RING and B-box 2 domains for the function of TRIMCyp and TRIM5αrh. Deletion of these domains from TRIM5αrh eliminated detectable restricting activity directed against HIV-1 and FIV, consistent with previous observations (Diaz-Griffero et al., 2007; Yap et al., 2007; Perez-Caballero et al., 2005; Javanbakht et al., 2005). By contrast, a TRIMCyp variant lacking the RING and B-box 2 domains exhibited partial ability to block HIV-1 and SIVagm infection. Moreover, RING- and B-box-2-deleted TRIMCyp fully restricted FIV. In a separate study (Javanbakht et al., in press), oligomerization of the cyclophilin A domain and L2 linker of TRIMCyp with a heterologous multimeric motif was shown to be sufficient to create an efficient HIV-1- and FIV-restricting factor. Cyclophilin A oligomerization results in increased avidity for the HIV-1 capsid (Diaz-Griffero et al., 2006b; Javanbakht et al., in press). These observations suggest a model in which increased cyclophilin A occupancy of the HIV-1 capsid results in inhibition of infection. The prolyl isomerase activity of the bound cyclophilin A may alter the conformation of the HIV-1 capsid protein and thereby modulate the uncoating process. TRIM5α, by contrast, has no such prolyl isomerase activity and thus may be more dependent on a B-box-2-directed function for restriction-related capsid uncoating. For example, the TRIM5α B-box 2 may recruit host cell uncoating factor(s) that contribute to restriction.
We also noted that FIV appears to be restricted by RING- and B-box-2-deleted TRIMCyp and artificially oligomerized cyclophilin A constructs more effectively than HIV- 1 or SIVagm. The FIV capsid may bind cyclophilin A more tightly, may intrinsically differ in stability from the HIV-1 and SIVagm capsids, or may be more susceptible to the effects of cyclophilin A binding or activity on the uncoating process. Future studies should clarify the basis for these differences in sensitivity to restriction.
HIV-1-infected cells treated with proteasome inhibitors exhibited large increases in the amount of particulate, cytosolic capsids, regardless of TRIM5αrh or TRIMCyp expression. Under these conditions, differences between the amounts of particulate capsids in the cytosol of cells expressing TRIM5αrh/TRIMCyp or control cells were obscured. Particulate HIV-1 capsids also persisted in the cytosol of cells expressing TRIMCyp mutants with altered B-box 2 domains, even though these mutants still inhibited HIV-1 infection. Thus, some level of restriction can be observed even when particulate capsid can still be detected in the cytosol of infected cells. This may reflect differences in the sensitivity of the infectivity and fate-of-capsid assays; for example, partially disrupted capsids may still pellet in the latter assay even though their infectivity is markedly compromised. Alternatively, other mechanisms of restriction may apply in the circumstance of proteasome inhibition, which results in major redistribution and coalescence of TRIM5α/TRIMCyp cytoplasmic aggregates. In future studies investigating these possibilities, it will be important to document the relevance of the restrictive mechanisms operative during proteasome inhibition to those pertaining to TRIM5α/TRIMCyp under natural conditions.
The anti-FIV potency of TRIM5αrh is dependent on variable region 1 of the B30.2 domain. This same region has been shown to contribute to the ability of TRIM5αrh to restrict HIV-1 and TRIM5αhu to restrict N-MLV (Stremlau et al., 2005; Perron, Stremlau, and Sodroski, 2006). The variable regions have been suggested to be surface-exposed loops on the B30.2(SPRY) domain, analogous to the complementarity-determining loops of immunoglobulins. Our results suggest that different TRIM5α proteins utilize common structural motifs for the recognition of diverse retroviral capsids. Future studies should shed light on the mechanistic details of TRIM5α-capsid interaction.
Materials and Methods
Plasmid construction
Plasmids expressing wild-type and mutant variants of TRIMCyp, TRIM5αrh and TRIM5αhu proteins tagged with a carboxy-terminal hemagglutinin (HA) epitope were constructed using PCR. The respective cDNA was PCR-amplified and digested with EcoRI and ClaI, whose sites were introduced by inclusion in each of the PCR primers. These fragments were cloned into the EcoRI and ClaI sites of the pLPCX vector (Stratagene). TRIM5αrh-TRIM5αhu chimeric proteins were named as previously described (Stremlau et al., 2005). The nomenclature for the chimerae is TRIM5 A(Bx-y), in which the encoded TRIM5 amino acids from x to y from species B are inserted into the TRIM5 protein of species A (H, human; R, rhesus monkey). The numbering scheme is based on the human TRIM5 residue numbers; the same numbers are used for therhesus monkey TRIM5 residues, based on the alignment of the TRIM5rh sequence to that of TRIM5hu.
Cells
Cf2Th canine thymocytes were obtained from ATCC. Cat CRFK cells were kindly provided by Dr. Eric Poeschla (Mayo Clinic).
Creation of cells stably expressing TRIMCyp and TRIM5α variants
Retroviral vectors encoding wild-type or mutant TRIMCyp or TRIM5α proteins were created using the pLPCX vector. Recombinant viruses were produced in 293T cells by cotransfecting the pLPCX plasmids with the pVPack-GP and pVPack-VSV-G packaging plasmids (Stratagene). The pVPack-VSV-G plasmid encodes the vesicular stomatitis virus (VSV) G envelope glycoprotein, which allows efficient entry into a wide range of vertebrate cells. Transduced cells were selected in 1 μg/ml puromycin (Sigma).
Infection with viruses expressing green fluorescent protein (GFP)
Recombinant HIV-1 and SIVagm expressing GFP were prepared as described (Stremlau et al., 2004). Viral stocks were quantified by measuring reverse transcriptase (RT) activity. For infections, 3 × 104 CRFK cells seeded in 24-well plates were incubated with virus for 24 hours. Cells were washed and returned to culture for 48 hours, and then subjected to FACS analysis with a FACScan (Becton Dickinson). The feline immunodeficiency virus (FIV) vector was obtained from System Biosciences (Mountain View, CA) and was prepared following the manufacturer’s instructions.
Protein analysis
Cellular proteins were extracted with radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris, pH 7.4; 100 mM NaCl; 1% sodium deoxycholate; 0.1% sodium dodecyl sulfate [SDS]; 1% NP-40; 2 mg/ml aprotinin; 2 mg/ml leupeptin; 1 mg/ml pepstatin A; 100 mg/ml phenylmethylsulfonyl fluoride). The cell lysates were analyzed by SDS-PAGE (10% acrylamide), followed by blotting onto nitrocellulose membranes (Amersham Pharmacia Biotech). Detection of protein by Western blotting utilized monoclonal antibodies directed against the HA (Roche) epitope tag, and monoclonal antibodies to β-actin (Sigma). Detection of proteins with HA epitope tags was performed by enhanced chemiluminescence (NEN Life Sciences Products), using an anti-rat-HRP secondary antibody (Amersham Pharmacia Biotech).
Fate-of-capsid assay
Cf2Th or CRFK cells were seeded on T80 plates at 20% confluence one day before the experiments. Cells were incubated with 10 ml of HIV-1 VLPs with or without envelope glycoproteins at 4°C for 30 minutes. The cultures were then shifted to 37°C and harvested at the indicated times. Samples were processed as previously described (Stremlau et al., 2006). Briefly, cells were lysed in a detergent-free butter. After low-speed centrifugation to pellet nuclei and cell debris, the cytosolic extracts “input” were layered onto a 50% sucrose cushion. After centrifugation, the “Pellet” was resuspended in 1X loading buffer and analyzed together with the “Input” by Western blot with a monoclonal antibody against the HIV-1 p24 capsid protein.
HIV CA-NC expression and purification
The HIV-1 CA-NC protein was expressed, purified and assembled as previously described (Ganser et al., 1999). The pET11a expression vector (Novagen) expressing CA-NC of HIV-1 was used to transform BL-21(DE3). CA-NC expression was induced with 1 mM isopropyl-beta-D-thiogalactopyranoside (IPTG) when cells achieved an optical density at 600 nm of 0.6. After 4 hours of induction, cells were harvested and resuspended in 20 mM Tris-HCl (pH 7.5), 1 μM ZnCl2, 10 mM 2-mercaptoethanol and protease inhibitors (Roche). Bacteria were lysed by sonication and debris was pelleted for 30 minutes at 35,000 × g. Nucleic acids were stripped from the solution by using 0.11 equivalents of 2M (NH4)2SO4 and the same volume of 10% polyethylenimine. Nucleic acids were removed by stirring and centrifugation at 29,500 × g for 15 minutes. Protein was recovered by addition of 0.35 equivalents of saturated (NH4)2SO4. Protein was centrifuged at 9,820 × g for 15 minutes and resuspended in 100 mM NaCl, 20 mM Tris-HCl (pH 7.5), 1 μM ZnCl2 and 10 mM 2-mercaptoethanol. Protein was dialyzed against 50 mM NaCl, 20 mM Tris-HCl (pH 7.5), 1 μM ZnCl2 and 10 mM 2-mercaptoethanol.
In vitro assembly of HIV-1 capsid complexes
HIV-1 capsid complexes were assembled in vitro by diluting the HIV-1 CA-NC protein to a concentration of 0.3 mM in 50 mM Tris–HCl (pH 8.0), 0.5 M NaCl and 2 mg/ml DNA oligo (TG)50. The mixture was incubated at 4°C overnight and centrifuged at 8,600 × g for 5 minutes. The pellet was resuspended in assembly buffer (50 mM Tris–HCl (pH 8.0), 0.5 M NaCl) at a final protein concentration of 0.15 mM (Ganser-Pornillos et al., 2004).
TRIM5 variants binding to HIV-1 CA-NC complexes
293T cells were transfected with plasmids expressing different TRIMCyp or TRIM5 proteins. Forty-eight hours after transfection, cell lysates were prepared as follows: Washed cells were resuspended in hypotonic lysis buffer (10 mM Tris pH 7.4, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT). The cell suspension was frozen and thawed, and then incubated on ice for 10 minutes. Afterwards, the lysate was centrifuged at 14,000 × g in a refrigerated microcentrifuge for 5 minutes. The supernatant was supplemented with 1/10 volume of 10X PBS and then used in the binding assay. Five ml of in vitro assembled CA particles were incubated with 200 μl of cell lysate at room temperature for 1 hour. A fraction of this mixture was stored (Input). The remainder of the mixture was spun through a 70% sucrose cushion (70% sucrose, 1X PBS and 0.5 mM DTT) at 100,000 × g in an SW55 rotor (Beckman) for 1 hour at 4°C. After centrifugation, the supernatant was carefully removed and the pellet was resuspended in 1X SDS-PAGE loading buffer (Pellet). The level of TRIM5 proteins was determined by Western blot.
Crosslinking of TRIM5 variants
The HA-tagged proteins were expressed transiently in 293T cells. Cells were washed in phosphate-buffered saline (PBS) and lysed in NP40 lysis buffer (0.5% Nonidet P40 (NP40), 1x protease inhibitor (complete EDTA-free, Roche Diagnostics) in PBS) for 45 minutes at 4°C. Lysates were centrifuged at 14,000 × g for 15 minutes at 4°C. The cleared lysates were not stored or frozen, but rather were directly crosslinked. Approximately 100–200 μl of cleared lysates were diluted with PBS/1 mM EDTA to a final volume of 400 μl. Lysates were crosslinked with varying concentrations (up to 10 mM) of ethylene glycol-bis(succinimidyl succinate) (EGS) for 30 minutes at room temperature and centrifuged briefly in a table-top centrifuge. The reaction mix was quenched with 0.1 M Tris– HCl, pH 7.5 and briefly centrifuged. The cleared, cross-linked lysates were precipitated with the anti-HA antibody HA.11 (Covance) and protein A-Sepharose beads (Amersham) for 2 hours at 4°C; final volumes for the immunoprecipitation were greater than 700 μl. The beads were washed four times with NP40 wash buffer (10 mM Tris–HCl, pH 7.5, 0.5 M NaCl, 0.5% NP40) and boiled in LDS sample buffer (106 mM Tris–HCl, 141 mM Tris Base, pH 8.5, 0.51 mM EDTA, 10% glycerol, 2% LDS, 0.22 mM SERVA Blue G250, 0.175 mM phenol red (Invitrogen)) with a final concentration of 1.2% β-mercaptoethanol (β-ME) for 10 minutes. Precipitated proteins were separated on 8% or 12% Tris–glycine gels, transferred to a PVDF membrane, and detected with the horseradish peroxidase-conjugated 3F10 anti-HA antibody (Roche Diagnostics) and the ECL Plus Western Blotting Detection System (Amersham).
Acknowledgments
We thank Ms. Yvette McLaughlin and Ms. Elizabeth Carpelan for manuscript preparation, and the National Institutes of Health (AI063987 and a Center for AIDS Research Award AI60354), the International AIDS Vaccine Initiative, the Bristol-Myers Squibb Foundation, and the late William F. McCarty-Cooper. M.S. was supported by a National Defense Science and Engineering Fellowship and was a fellow of the Ryan Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson JL, Campbell EM, Wu X, Vandegraaff N, Engelman A, Hope TJ. Proteasome inhibition reveals that a functional preintegration complex intermediate can be generated during restriction by diverse TRIM5 proteins. J Virol. 2006;80(19):9754–60. doi: 10.1128/JVI.01052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan S, Hatziioannou T, Cunningham T, Muesing MA, Gottlinger HG, Bieniasz PD. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc Natl Acad Sci U S A. 2002;99(18):11914–9. doi: 10.1073/pnas.162299499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Griffero F, Li X, Javanbakht H, Song B, Welikala S, Stremlau M, Sodroski J. Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology. 2006a;349:300–15. doi: 10.1016/j.virol.2005.12.040. [DOI] [PubMed] [Google Scholar]
- Diaz-Griffero F, Vandegraaff N, Li Y, McGee-Estrada K, Stremlau M, Welikala S, Si Z, Engelman A, Sodroski J. Requirements for capsid-binding and an effector function in TRIMCyp-mediated restriction of HIV-1. Virology. 2006b;351(2):404–19. doi: 10.1016/j.virol.2006.03.023. [DOI] [PubMed] [Google Scholar]
- Diaz-Griffero F, Kar A, Perron M, Xiang SH, Javanbakht H, Li X, Sodroski J. Modulation of retroviral restriction and proteasome inhibitor-resistant turnover by changes in the TRIM{alpha} B-box 2 domain. J Virol Jul. 2007;11 doi: 10.1128/JVI.00703-07. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble TR, Vajdos FF, Yoo S, Worthylake DK, Houseweart M, Sundquist WI, Hill CP. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87(7):1285–94. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283(5398):80–3. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- Ganser-Pornillos BK, von Schwedler UK, Stray KM, Aiken C, Sundquist WI. Assembly properties of the human immunodeficiency virus type 1 CA protein. J Virol. 2004;78(5):2545–52. doi: 10.1128/JVI.78.5.2545-2552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatziioannou T, Perez-Caballero D, Cowan S, Bieniasz PD. Cyclophilin interactions with incoming human immunodeficiency virus type 1 capsids with opposing effects on infectivity in human cells. J Virol. 2005;79(1):176–83. doi: 10.1128/JVI.79.1.176-183.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatziioannou T, Perez-Caballero D, Yang A, Cowan S, Bieniasz PD. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5alpha. Proc Natl Acad Sci U S A. 2004;101(29):10774–9. doi: 10.1073/pnas.0402361101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann W, Schubert D, LaBonte J, Munson L, Gibson S, Scammell J, Ferrigno P, Sodroski J. Species-specific, postentry barriers to primate immunodeficiency virus infection. J Virol. 1999;73(12):10020–8. doi: 10.1128/jvi.73.12.10020-10028.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javanbakht H, Diaz-Griffero F, Stremlau M, Si Z, Sodroski J. The contribution of RING and B-box 2 domain in retroviral restriction mediated by monkey TRIM5alpha. J Biol Chem. 2005;29:26933–40. doi: 10.1074/jbc.M502145200. [DOI] [PubMed] [Google Scholar]
- Javanbakht H, Yuan W, Yeung DF, Song B, Diaz-Griffero F, Li Y, Li X, Stremlau M, Sodroski J. Characterization of TRIM5alpha trimerization and its contribution to human immunodeficiency virus capsid binding. Virology. 2006;353(1):234–46. doi: 10.1016/j.virol.2006.05.017. [DOI] [PubMed] [Google Scholar]
- Javanbakht H, Diaz-Griffero F, Yuan W, Yeung DF, Li X, Song B, Sodroski J. The ability of multimerized cyclophilin A to restrict retrovirus infection. Virology. doi: 10.1016/j.virol.2007.04.034. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keckesova Z, Ylinen LM, Towers GJ. The human and African green monkey TRIM5alpha genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc Natl Acad Sci U S A. 2004;101(29):10780–5. doi: 10.1073/pnas.0402474101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li X, Stremlau M, Lee M, Sodroski J. Removal of arginine 332 allows human TRIM5alpha to bind human immunodeficiency virus capsids and to restrict infection. J Virol. 2006;80(14):6738–44. doi: 10.1128/JVI.00270-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TY, Emerman M. Cyclophilin A interacts with diverse lentiviral capsids. Retrovirology. 2006;3:70. doi: 10.1186/1742-4690-3-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luban J, Bossolt KL, Franke EK, Kalpana GV, Goff SP. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell. 1993;73(6):1067–78. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- Mische CC, Javanbakht H, Song B, Diaz-Griffero F, Stremlau M, Strack B, Si Z, Sodroski J. Retroviral restriction factor TRIM5alpha is a trimer. J Virol. 2005;22:14464–50. doi: 10.1128/JVI.79.22.14446-14450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munk C, Brandt SM, Lucero G, Landau NR. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc Natl Acad Sci U S A. 2002;99(21):13843–8. doi: 10.1073/pnas.212400099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisole S, Lynch C, Stoye JP, Yap MW. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc Natl Acad Sci U S A. 2004;101(36):13324–8. doi: 10.1073/pnas.0404640101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JC. Gene transfer vectors derived from equine infectious anemia virus. Gene Ther. 1998;5(11):1481–7. doi: 10.1038/sj.gt.3300768. [DOI] [PubMed] [Google Scholar]
- Owens CM, Song B, Perron MJ, Yang PC, Stremlau M, Sodroski J. Binding and susceptibility to postentry restriction factors in monkey cells are specified by distinct regions of the human immunodeficiency virus type 1 capsid. J Virol. 2004;78(10):5423–37. doi: 10.1128/JVI.78.10.5423-5437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens CM, Yang PC, Gottlinger H, Sodroski J. Human and simian immunodeficiency virus capsid proteins are major viral determinants of early, postentry replication blocks in simian cells. J Virol. 2003;77(1):726–31. doi: 10.1128/JVI.77.1.726-731.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Caballero D, Hatziioannou T, Yang A, Cowan S, Bieniasz PD. Human tripartite motif 5alpha domains responsible for retrovirus restriction activity and specificity. J Virol. 2005a;79(14):8969–78. doi: 10.1128/JVI.79.14.8969-8978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Caballero D, Hatziioannou T, Zhang F, Cowan S, Bieniasz PD. Restriction of human immunodeficiency virus type 1 by TRIM-CypA occurs with rapid kinetics and independently of cytoplasmic bodies, ubiquitin, and proteasome activity. J Virol. 2005b;79(24):15567–72. doi: 10.1128/JVI.79.24.15567-15572.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron MJ, Stremlau M, Lee M, Javanbakht H, Song B, Sodroski J. The human TRIM5alpha restriction factor mediates accelerated uncoating of the N-tropic murine leukemia virus capsid. J Virol. 2007;81(5):2138–48. doi: 10.1128/JVI.02318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron MJ, Stremlau M, Sodroski J. Two surface-exposed elements of the B30.2/SPRY domain as potency determinants of N-tropic murine leukemia virus restriction by human TRIM5alpha. J Virol. 2006;80(11):5631–6. doi: 10.1128/JVI.00219-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JP, Menezes AN, Moreira MA, Bonvicino CR, Seuanez HN, Soares MA. Evolution of cyclophilin A and TRIMCyp retro-transposition in New World primates. J Virol. 2005;79(23):14998–03. doi: 10.1128/JVI.79.23.14998-15003.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz DT, Teo W, Olsen JC, Poeschla EM. Restriction of feline immunodeficiency virus by Ref1, Lv1, and primate TRIM5alpha proteins. J Virol. 2005;79(24):15175–88. doi: 10.1128/JVI.79.24.15175-15188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci U S A. 2005;102(8):2832–7. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430(6999):569–73. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- Song B, Gold B, O’Huigin C, Javanbakht H, Li X, Stremlau M, Winkler C, Dean M, Sodroski J. The B30.2(SPRY) domain of the retroviral restriction factor TRIM5alpha exhibits lineage-specific length and sequence variation in primates. J Virol. 2005a;79(10):6111–21. doi: 10.1128/JVI.79.10.6111-6121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B, Javanbakht H, Perron M, Park DH, Stremlau M, Sodroski J. Retrovirus restriction by TRIM5alpha variants from Old World and New World primates. J Virol. 2005b;79(7):3930–7. doi: 10.1128/JVI.79.7.3930-3937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427(6977):848–53. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci U S A. 2006;103(14):5514–9. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, Perron M, Welikala S, Sodroski J. Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J Virol. 2005;79(5):3139–45. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers GJ, Hatziioannou T, Cowan S, Goff SP, Luban J, Bieniasz PD. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat Med. 2003;9(9):1138–43. doi: 10.1038/nm910. [DOI] [PubMed] [Google Scholar]
- Wu X, Anderson JL, Campbell EM, Joseph AM, Hope TJ. Proteasome inhibitors uncouple rhesus TRIM5alpha restriction of HIV-1 reverse transcription and infection. Proc Natl Acad Sci U S A. 2006;103(109):7465–70. doi: 10.1073/pnas.0510483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap MW, Nisole S, Stoye JP. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr Biol. 2005;15(1):73–8. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- Yap MW, Mortuza GB, Taylor IA, Stoye JP. The design of artificial retroviral restriction factors. Virology. 2007 May 8; doi: 10.1016/j.virol.2007.04.005. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Yin L, Braaten D, Luban J. Human immunodeficiency virus type 1 replication is modulated by host cyclophilin A expression levels. J Virol. 1998;72(8):6430–6. doi: 10.1128/jvi.72.8.6430-6436.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]


