Abstract
The invasion of human malignant melanoma cells into the extracellular matrix (ECM) involves the accumulation of proteases at sites of ECM degradation where activation of matrix metalloproteases (MMP) occurs. Here, we show that when membrane type 1 MMP (MT-MMP) was overexpressed in RPMI7951 human melanoma cells, the cells made contact with the ECM, activated soluble and ECM-bound MMP-2, and degraded and invaded the ECM. Further experiments demonstrated the importance of localization of the MT-MMP to invadopodia. Overexpression of MT-MMP without invadopodial localization caused activation of soluble MMP-2, but did not facilitate ECM degradation or cell invasiveness. Up-regulation of endogenous MT-MMP with concanavalin A caused activation of MMP-2. However, concanavalin A treatment prevented invadopodial localization of MT-MMP and ECM degradation. Neither a truncated MT-MMP mutant lacking transmembrane (TM) and cytoplasmic domains (ΔTMMT-MMP), nor a chimeric MT-MMP containing the interleukin 2 receptor α chain (IL-2R) TM and cytoplasmic domains (ΔTMMT-MMP/TMIL-2R) were localized to invadopodia or exhibited ECM degradation. Furthermore, a chimera of the TM/cytoplasmic domain of MT-MMP (TMMT-MMP) with tissue inhibitor of MMP 1 (TIMP-1/TMMT-MMP) directed the TIMP-1 molecule to invadopodia. Thus, the MT-MMP TM/cytoplasmic domain mediates the spatial organization of MT-MMP into invadopodia and subsequent degradation of the ECM.
Cell invasion through the extracellular matrix (ECM) is a driving force for tissue development and cancer metastasis (1, 2). Invading cells possess ECM degrading proteolytic enzymes that appear at unique surface structures, which we have termed invadopodia (3). The factors that promote protease localization at sites of cell invasion are not known. However, increasing evidence indicates that tumor cell invasion involves the accumulation of membrane-bound proteases that direct activation of workhorse enzymes such as the 72-kDa matrix metalloprotease (MMP), MMP-2, on the cell surface (2–5). Surface activation of MMP-2 can be stimulated by cultivation of cells on collagen (6) or gelatin (7), or treatment with concanavalin A (Con A) (8). Such activation is sensitive to synthetic MMP inhibitors (9–11) and tissue inhibitor of MMP 2 (TIMP-2) (12). Recent studies showed that MMP-2 activation is mediated by membrane type 1 MMP (MT-MMP) in the capacity of either an activator (12–14) or a receptor (15). MT-MMP is constitutively activated by a furin-like pathway, and may also directly degrade ECM (16–18). Thus, it is possible that MT-MMP functions both in MMP-2 activation, and in localizing such proteases to sites of cell invasion.
A limitation of biochemical analysis of MMP-2 activation by MT-MMP is that the degradative and invasive activities of the cell can be compared only to the relative amount of proteases or activators present, whereas its correlation with the most relevant parameter of cell invasion—the cellular site of protease activation and action—remains unknown. Cell surface proteolytic activity has been identified at specialized membrane extensions, invadopodia, of a moderately invasive RPMI-7951 human melanoma cell line (19). We have expressed full-length and mutant MT-MMP cDNAs in RPMI-7951 cells and examined the activation of soluble and ECM-bound MMP-2 as well as the degradative and invasive activities of the transfected cells. Proteolytic activity of soluble and ECM-bound MMP were measured to assess sites of protease action. We show here that overexpression of the membrane-bound protease activator MT-MMP resulted in localization of proteases at invadopodia, and initiated a proteolytic cascade for cell invasion. Furthermore, we demonstrated that the transmembrane (TM)/cytoplasmic domain of MT-MMP (TMMT-MMP, 542VVLPVLLLLLVLAVGLAVFFFRRHGTPRRLLYCQRSLLDKV582) was required for invadopodial localization of the enzyme and therefore for directing cellular invasion, as neither occurred with MT-MMP mutants lacking the TMMT-MMP (ΔTMMT-MMP) or containing the interleukin 2 receptor α chain (IL-2R) TMIL-2R (ΔTMMT-MMP/TMIL-2R).
MATERIALS AND METHODS
Materials.
The human melanotic melanoma RPMI-7951 (ATCC HTB 66) was purchased from the American Type Culture Collection. Mouse mAb 113–5B7 directed against MT-MMP (13) was kindly provided by K. Iwata (Fuji). Mouse antibody 7–6C1 against TIMP-1 was purchased from Oncogene Science. The metalloprotease inhibitor CT1847 (20) was kindly provided by A. Docherty (Celltech, Slough, U.K.).
Cell Culture and in Vitro Degradation/Invasion Assays.
RPMI-7951 cells were cultured in a 1:1 mixture of DMEM and RPMI medium 1640 supplemented with 10% calf serum, 5% Nu-serum (Collaborative Research), 2 mM l-glutamine, 1 unit/ml penicillin, and 10 μg/ml streptomycin as described (21). RPMI-7951 is a moderately invasive human melanoma cell line, secreting 72 kDa MMP-2, which converts to 64- and 62-kDa forms when activated, and expressing background level of membrane proteases (19). For the fibronectin degradation/invasion assay, cells transfected with MT-MMP and its mutants were collected by trypsinization and 105 cells were added to fluorescein-fibronectin-coated crosslinked gelatin films on a 15-mm round glass coverslip in a 12-well plate as described (19, 22). This technique measures the fibronectin degradation by cells as well as the invasiveness in terms of foci of invadopodial extensions and surface indentations in the crosslinked gelatin film. For the former, cells were allowed to grow on the films for the indicated times, photographed using the Planapo 63/1.4 or 25/1.2 objectives on a Zeiss Photomicroscope III under epifluorescence, and digitized from photographic negatives using Optimas image analyzer coupled to a Panasonic WV-CD50 camera. For the latter, cells that had been cultured on fibronectin-gelatin films for 12 h were analyzed for the appearance of invadopodial extensions into the crosslinked gelatin film using Nomarski Differential Contrast microscopy as described (19). In the case of treatment with Con A (25 μg/ml) (8) or protease inhibitors, cells were allowed to attach to fluorescent fibronectin-coated, crosslinked gelatin films in complete medium at 37°C for 1 h, and then incubated in experimental conditions (serum-free DMEM, Con A, or inhibitors indicated in the figure legends), for additional 5 h. For visualizing MT-MMP or TIMP-1 localization, transfected cells were allowed to spread on fluorescein-fibronectin-coated crosslinked gelatin films for 3 h, and stained with mouse mAb 113–5B7 against MT-MMP or mouse mAb 7–6C1 against TIMP-1 (Oncogene Science) at 37°C for 30 min, fixed with cold 95% acetone for 5 min, and developed with tetramethylrhodamine B isothiocyanate-goat anti-mouse IgG (10 μg/ml; Cappel) as described (23).
Vector Construction and Transfection of RPMI-7951 Cells.
The pSG5 plasmids for expression of wild-type MT-MMP, ΔTMMT-MMP, and the chimera: ΔTMMT-MMP/TMIL-2R, TIMP-1/TMMT-MMP, and TIMP-1/TMIL-2R, were constructed by manipulating the cDNAs of these proteins as described (23). Cells were transfected with pSG5 plasmid lacking an insert (Mock) or containing sequences encoding MT-MMP or other mutant proteins above using LipofectAMINE method (Life Technologies, Grand Island, NY). Transfected cells were cultured for 48 h and suspended by trypsin/EDTA for in vitro degradation/invasion assay or immunofluorescence as described above. To verify that MT-MMP or recombinant proteins were expressed in transfected cells, cells were examined by indirect immunofluorescence staining or by a combination of cell surface biotinylation and immunoprecipitation as described (23). Among these proteins, only ΔTMMT-MMP was detected in the culture medium conditioned by ΔTMMT-MMP-transfected cells, whereas MT-MMP and other chimeric proteins were localized on the cell surface.
Subcellular Fractionation.
Fractionation was performed as described (24). Cell bodies were sheared from adherent cells using an “L” shaped Pasteur pipette. Proteins associated with invadopodia and the ECM were extracted by collecting the gelatin substratum and solubilizing proteins within it using RIPA buffer (1% Nonidet P-40/0.5% sodium deoxycholate/0.1% SDS in TBS, pH 8.0.).
Gelatin Zymography for Visualizing MMP-2 Activation.
The serum-free medium conditioned by transfected cells, or cells treated with Con A or protease inhibitors, was collected from ≈106 cells after 2 days of growth. Conditioned medium was centrifuged (1,000 × g for 10 min) to remove any cellular debris. Aliquots from the medium, or cell lysate or invadopodia/ECM extract described above were solubilized in SDS/PAGE sample buffer and analyzed by gelatin zymography as described (8).
RESULTS
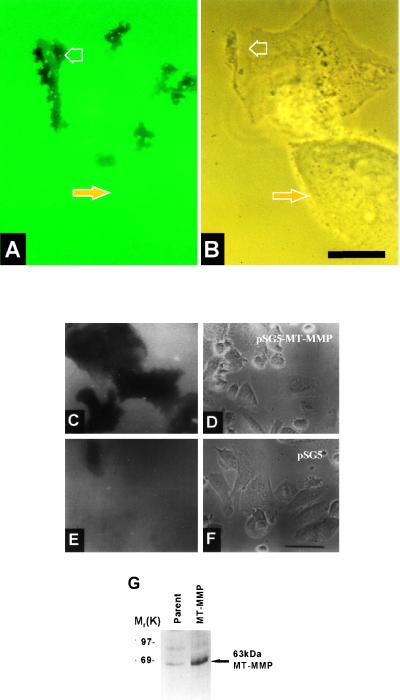
To investigate the biological consequences of MT-MMP expression, RPMI-7951 cells were transfected with MT-MMP or a vector control, collected by trypsinization and plated on fluorescein-fibronectin-coated crosslinked gelatin films. Within 1–3 h, cells transfected with full-length MT-MMP initiate localized fibronectin/gelatin degradation in association with invadopodia at the leading edge of the cell (Fig. 1 A and B; open arrow). Nontransfected cells show very little fibronectin/gelatin degradation (Fig. 1 A and B; solid arrow). Cells overexpressing MT-MMP degraded the area of film in contact with the cells within 6 h of seeding on fibronectin/gelatin films (Fig. 1 C and D) whereas nontransfected cells and controls did not (Fig. 1 E and F). To confirm MT-MMP expression by transfected cells, mAb 113–5B7 was used to immunoprecipitate proteins from surface biotinylated cells. A 63-kDa protein, the expected size for mature MT-MMP, was immunoprecipitated from a lysate of RPMI-7951 cells transfected with MT-MMP, but very little protein was immunoprecipitated from the parental cell line lysate (Fig. 1G). These results demonstrate that MT-MMP is a key cell surface protease involved in degradation of the ECM at invadopodia.
Figure 1.
Degradation/invasion of fibronectin-coated crosslinked gelatin substrata by melanoma cells overexpressing MT-MMP. RPMI-7951 cells transfected with MT-MMP were plated onto fluorescein-fibronectin coated gelatin films. After 1 h cells were photographed under epifluorescence (A) or by phase contrast microscopy (B). Open arrows indicate degradation spots (A) that correspond to invadopodial complexes at the leading edge of the transfected cell (B). The solid arrows mark a nontransfected cell in the same field that has not produced any degradation spots. RPMI-7951 cells transfected with pSG5 containing MT-MMP (C and D) or with pSG5 alone (E and F) were plated onto fluorescein-fibronectin coated gelatin films. After 6 h cells were photographed under epifluoresence (C and E) to reveal fluorescein-fibronectin degradation or by phase contrast microscopy (D and F). Cells overexpressing MT-MMP extensively degraded the area of film in contact with the cells (C and D) whereas cells transfected with vector alone did not (E and F). Bars: A and B, 10 μm; C–F, 50 μm; G, RPMI-7951 cells transfected with MT-MMP and parental cells were surface-labeled with biotin. Proteins were immunoprecipitated from cell lysates using mAb 113–5B7 against MT-MMP then detected using anti-biotin horse radish peroxidase.
To determine whether activation of MMP-2 can lead to matrix degradation, RPMI-7951 cells transfected with MT-MMP or treated with Con A were analyzed for gelatinase activation, fibronectin/gelatin degradation, and invasion. Conditioned medium and cell lysate from cells transfected with full-length MT-MMP or treated with Con A contained the 64- and 62-kDa active forms of MMP-2, while that of parental and mock transfected cells contained the latent 72-kDa form (Fig. 2A). The activation mediated by MT-MMP transfection was blocked by the synthetic matrix metalloprotease inhibitor CT1847, and was not sensitive to inhibitors of serine, cysteine, or aspartate proteases (Fig. 2B). RPMI-7951 cells overexpressing MT-MMP degraded more fibronectin/gelatin film than their counterparts transfected with vector alone (mock cells), parental cells treated with Con A (Fig. 2C), or MT-MMP expressing cells treated with Con A (Fig. 2D). In 6–12 h, mock-transfected cells exhibited similar fibronectin/gelatin degradation patterns to Con A treated cells, i.e., very few degradation spots (Fig. 2 C and D). In contrast, cells expressing MT-MMP began to degrade the fibronectin/gelatin substratum within 3 h of plating, and digested most of the fibronectin/gelatin after 12 h (Fig. 2C, MT-MMP). When this increase in fibronectin degradation was quantified, we found that significantly more fibronectin substratum (ρ < 1 × 10−5) was removed by MT-MMP transfected cells compared with mock cells, or either mock or MT-MMP transfected cells treated with Con A (Fig. 2D). The enhanced fibronectin degradation by MT-MMP transfected cells paralleled their subsequent increase in invasion into the crosslinked gelatin film (Fig. 2E), as measured by percentage of cells extending their invadopodia into the film (ρ < 1 × 10−5). Consistent with the inhibition of MMP-2 activation by the metalloprotease inhibitor CT1847 (Fig. 2B), the fibronectin degradation by MT-MMP overexpressing cells was also inhibited by CT1847 but not by other protease inhibitors (Fig. 2F). These data suggest that MMP-2 activation is necessary but not sufficient for localized fibronectin degradation and directed invasion.
Figure 2.
Activation of MMP-2 and degradation/invasion of fibronectin-coated crosslinked gelatin substrata by melanoma cells treated with Con A or overexpressing MT-MMP. (A) Activity of MMP-2 released into the medium (Medium) or associated with cells (Cell lysate) in the presence (Con A) or absence (Parent) of Con A or transfected with pSG5-MT-MMP (MT-MMP) were analyzed by gelatin zymography. The three bands shown represent 72-kDa pro-MMP-2, the 62-kDa intermediate form, and the 59-kDa active form of MMP-2. (B) Activity of MMP-2 released into the medium (Conditioned medium) by the cells or associated with cells transfected with pSG5-MT-MMP (Cell lysate) in the absence (MT-MMP) or presence of the following protease inhibitors: 100 nM CT1847 (InMMP), 200 nM CT1847 (InMMP′), 320 μg/ml ɛ-amino caproic acid (ACA), 5 μg/ml pepstatin A (PSA), and 10 μg/ml leupeptin (LP). Gelatinase activities were analyzed by gelatin zymography. (C) Fibronectin degradation by cells transfected with pSG5 plasmid (Mock), treated with Con A, or transfected with pSG5-MT-MMP (MT-MMP). Cells were allowed to spread on fluorescein-fibronectin-coated crosslinked gelatin films and were photographed with fluorescence microscopy after 1, 3, 6, and 12 h. Wide black areas in the lower panels indicate the degradation of fibronectin-gelatin films by cells. Mock transfectants and the cells treated with Con A showed very little black area on the film, indicating background levels of fibronectin degradation by the melanoma cells, whereas cells transfected with MT-MMP show much higher levels of degradation. Bar = 25 μm. (D) The areas of degraded fibronectin substratum under the cells transfected with pSG5 plasmid (Mock) or transfected with pSG5-MT-MMP (MT-MMP) in the absence or presence of Con A were measured with Optimas image analyzer after 6 h. Each value represents the mean of three separate determinations (among 100 cells) ± SD. Duplicate experiments gave similar results. (E) The invasiveness of the cells transfected with pSG5 plasmid (Mock) or transfected with pSG5-MT-MMP (MT-MMP) in the absence or presence of Con A after 12 h was analyzed in terms of percentage of cells forming invadopodial extensions in the crosslinked gelatin film. Each value represents the mean of three separate determinations (among 100 cells) ± SD. (F) The areas of degraded fibronectin substratum under the cells in the presence of protease inhibitors as indicated above were measured with Optimas image analyzer. Each value represents the mean of three separate determinations (among 100 cells) ± SD.
A previous study has shown that the TMMT-MMP could function as a membrane linker when fused to TIMP-1, and that a chimera of MT-MMP and the TMIL-2R was retained on the cell surface and activated MMP-2 (23). We tested here the role of the TMMT-MMP in localization to invadopodia and in the degradation/invasion phenotype of melanoma cells. During cell invasion, structures described as “invadopodial complexes” are formed (25). Such structures appear to represent immature invadopodia at the leading edge of the cell in close contact with the matrix (Fig. 1 A and B), and they extend to form the invasion front of the cell. Immunofluorescent staining using mAb 113–5B7 shows that MT-MMP is predominantly associated with invadopodial complexes at the leading edge of a transfected cell (Fig. 3A), corresponding to the areas where fibronectin/gelatin degradation occurs (Fig. 3B). Cells overexpressing full-length MT-MMP localized the molecule at invadopodial complexes (Fig. 3C) as demonstrated by colocalization with the invadopodial marker A27 (25) (Fig. 3D). However cells transfected with vector pSG5 showed no such invadopodial MT-MMP staining (data not shown). Con A treatment of MT-MMP transfected RPMI-7951 cells significantly reduced their ability to degrade fibronectin/gelatin and their invasive behavior (Fig. 2 C–E), but did not affect their ability to activate MMP-2 (Fig. 2A). Immunostaining showed that Con A treatment abolished MT-MMP localization to invadopodia as MT-MMP was aggregated on cell surfaces other than A27 labeled invadopodia (Fig. 3 E and F). To assess the role of various domains of MT-MMP in the invadopodial localization, we used a variety of deletion mutants as well as chimeras formed between TMMT-MMP or TMIL-2R and the extracellular domain of MT-MMP or a secreted protein, TIMP-1. Invadopodial localization was seen in a chimeric TIMP-1 mutant with the TMMT-MMP (Fig. 3G). However, invadopodial localization of MT-MMP was abolished by truncation of the TM domain (data not shown), and was not seen in a chimera of ΔTMMT-MMP and the TMIL-2R (Fig. 3 H). These results show that the TMMT-MMP is critical for mediating invadopodial localization.
Figure 3.
The TM/cytoplasmic domain is required to localize MT-MMP at invadopodial complexes. Arrows in individual panels indicate sites of invadopodial complexes. (A and B) RPMI-7951 cells transfected with MT-MMP were plated on a fluorescein-fibronectin crosslinked gelatin film. Cells were stained for MT-MMP using mAb 113–5B7 (A) and areas of fibronectin degradation were visualized by epifluorescence (B). (C and D) RPMI-7951 cells transfected with MT-MMP were cultured on fibronectin-gelatin films for 3 h, and double immunostained with mAb113–5B7 against MT-MMP (C) and antibody A27 against invadopodia (D). Double labeling of MT-MMP transfected cells shows that MT-MMP (C) colocalizes with invadopodia as labeled by mAb A27 (D). (E and F) RPMI-7951 cells transfected with MT-MMP were cultured on fibronectin-gelatin films in the presence of Con A (25 μg/ml) for 3 h, and double immunostained with mAb113–5B7 against MT-MMP (E) and antibody A27 against invadopodia (F). MT-MMP loses its invadopodial localization in the presence of Con A. (G) RPMI-7951 cells transfected with TIMP-1/TMMT-MMP were cultured on fibronectin-gelatin films for 3 h, and immunostained with antibody 7–6C1 against TIMP-1. A cell expressing TIMP-1/TMMT-MMP localized this chimera at invadopodial complexes as revealed by labeling with an anti-TIMP-1 antibody (G). (H) RPMI-7951 cells transfected with chimeric ΔTMMT-MMP/TMIL-2R were cultured on fibronectin-gelatin films for 3 h, and immunostained with mAb113–5B7 against MT-MMP. A cell expressing ΔTMMT-MMP/TMIL-2R failed to localize MT-MMP to invadopodia (H) as shown by staining with mAb 113–5B7.
Similar to cells treated with Con A or transfected with MT-MMP, the cells transfected with a chimeric ΔTMMT-MMP/TMIL-2R mediated activation of MMP-2 into the medium (Fig. 4A). Cells overexpressing MT-MMP had active MMP-2 associated with invadopodial membranes and the ECM (Fig. 4B). In contrast cells expressing either ΔTMMT-MMP or ΔTMMT-MMP/TMIL-2R failed to activate ECM or invadopodia-associated MMP-2 (Fig. 4B), degrade the fibronectin substratum (Fig. 4C) or to invade the matrix (Fig. 4D). Mock-transfected cells and cells transfected with ΔTMMT-MMP, TIMP-1, chimeric TIMP-1/TMIL-2R, or chimeric TIMP-1/TMMT-MMP did not activate MMP-2 or degrade/invade matrix (Fig. 4 A–D). The fibronectin degradation/invasion function of MT-MMP was not seen in cells expressing ΔTMMT-MMP or chimeric ΔTMMT-MMP/TMIL-2R (Fig. 4 C and D), although these MT-MMP mutants were still retained on the cell surface, and the latter could activate MMP-2 in the medium (Fig. 4A). Thus, the TM/cytoplasmic and catalytic domains of MT-MMP are required for the activation of ECM-bound gelatinase, local ECM degradation and subsequent invasion of melanoma cells.
Figure 4.
The TM/cytoplasmic and catalytic domains of MT-MMP are required for activation of ECM-bound MMP-2, local ECM degradation and subsequent invasion by melanoma cells. Gelatinase activities of MMP-2 released into the medium by the cells transfected with pSG5 lacking an insert (Mock), pSG5 plasmids containing MT-MMP (MT-MMP), truncated mutant lacking the TM domain (ΔTMMT-MMP), chimeric ΔTMMT-MMP/TMIL-2R, TIMP-1, chimeric TIMP-1/TMIL-2R, or chimeric TIMP-1/TMMT-MMP, were analyzed by gelatin zymography. (B) Gelatinase activities of MMP-2 associated with the ECM and invadopodial membranes of cells transfected with chimeric MT-MMP molecules. Cells cultured on crosslinked gelatin films were fractionated as described (24) and ECM/invadopodial fractions were examined by gelatin zymography. (C) The fibronectin-degrading activities of transfectants were measured with Optimas image analyzer after 6 h. Each value represents the mean of three separate determinations (among 100 cells) ± SD. (D) The invasiveness of transfectants after 12 h was analyzed in terms of the percentage of cells forming invadopodial extensions in the crosslinked gelatin film. Each value represents the mean of three separate determinations (among 100 cells) ± SD.
DISCUSSION
We have shown that overexpression of MT-MMP increases RPMI-7951 human melanoma cell invasion by stimulating invadopodial activity. The membrane-bound protease localizes predominantly to invadopodia that are sites of cellular ECM degradation. Thus, MT-MMP may participate a protease cascade involving enzymes such as seprase and MMP-2 (19, 26–28) that leads to an increase in melanoma cell invasiveness. We also showed that the TM/cytoplasmic domain of MT-MMP mediates the spatial organization of MT-MMP to invadopodia and that this is necessary for degradation of the ECM and subsequent invasion. Our data suggest a membrane docking mechanism for melanoma cell invasion in which cell invasiveness can be initiated when active proteases are recruited to sites of ECM degradation.
The biochemical mechanisms by which MMP-2 is involved in invasion are not well-understood. It is known that the secreted MMP-2 can be recruited to tumor cell invadopodia and that it can bind to the ECM (6, 7, 19, 29–31). RPMI-7951 cells produce MMP-2 when cultured on the crosslinked gelatin film, and ELISA analysis showed that up to 80% of the proenzyme became bound to crosslinked gelatin films (19). Also, we showed that active MMP-2 was localized on invadopodia while other matrix-bound and secreted MMP-2 remained latent (6, 19). In this report, we showed that MT-MMP expression can induce activation of both soluble and ECM-bound MMP-2 at sites of invadopodial contact. However, secreted MMP-2 can be activated by cells expressing MT-MMP or ΔTMMT-MMP/TMIL-2R, and by cells treated with Con A (Fig. 2A and 4A). Con A treatment significantly (ρ < 1 × 10−5) inhibited the fibronectin/gelatin degradation (Fig. 2D) and invasion (Fig. 2E) of the cells expressing MT-MMP. This maybe explained by the fact that Con A treatment prevents MT-MMP from localizing to invadopodia (Fig. 3 E and F). Con A may crosslink membrane glycoproteins, including MT-MMP, on available dorsal cell surfaces, thus the activation of soluble MMP-2 can proceed but localization of MT-MMP to invadopodial complexes is prevented.
In addition to activating MMP-2 (12–14), MT-MMP can serve as a MMP-2 receptor (15) and thus could be involved in the recruitment of the MT-MMP/MMP-2 complex to sites of invasion. Recent studies showed that MT-MMP may also directly degrade ECM components including fibronectin, collagen and gelatin (16–18). Thus, MT-MMP may play dual roles in degrading ECM and in activating matrix-bound proenzyme at invadopodial sites to extend the proteolytic cascade.
RPMI-7951 cells transfected with ΔTMMT-MMP or chimeric ΔTMMT-MMP/TMIL-2R failed to localize MT-MMP at invadopodia, and were not capable of activating matrix-bound MMP-2, degrading ECM, or invading the matrix. Both MT-MMP and TIMP-1/TMMT-MMP localized to invadopodia (Fig. 3 C and G), but only the former coincided with the local fibronectin/gelatin degradation (Fig. 3 A and B). These data suggest a model where fibronectin degradation by an invading cell is due to the binding of the TM/cytoplasmic domain of MT-MMP to an as yet unidentified docking system, resulting in recruitment of the protease into invadopodial complexes. Such colocalization of proteases and matrix substrates will then result in a degradative interaction. We propose that the TM/cytoplasmic domain of MT-MMP contains specific sequences that are involved in the localization of these proteases to invadopodia contacting the ECM, thus providing a spatially specific mechanism for the regulation of MMP-2 activation, matrix degradation and cellular invasion into the ECM.
Acknowledgments
We are most grateful to K. Iwata (Fuji Chemical Industries, Ltd., Takaoka, Japan) for providing mouse mAb 113–5B7; A. Docherty (Celltech, Slough, U.K.) for metalloprotease inhibitors; and D. Flessate, L. A. Goldstein, S. C. Mueller, M. L. Piñeiro-Sánchez (Georgetown University, Washington, DC), and Y. Yamada (National Institute of Dental Research, National Institutes of Health) for critical discussion. This work was supported by U.S. Public Health Service Grants R01 CA-39077 and HL-33711 to W.-T.C., and in part by U.S. Public Health Service Grant 2P30-CA-51008, the Lombardi Cancer Center Microscopy, and Imaging Shared Resource.
ABBREVIATIONS
- Con A
concanavalin A
- ECM
extracellular matrix
- IL-2R
interleukin 2 receptor α chain
- MMP
matrix metalloprotease
- MT-MMP
membrane type 1 MMP
- TIMP-1 and -2
tissue inhibitor of MMP-1 and -2
- TM
transmembrane domain
- TMMT-MMP
TM/cytoplasmic domain of MT-MMP
References
- 1.Mignatti P, Rifkin D B. Physiol Rev. 1993;73:161–195. doi: 10.1152/physrev.1993.73.1.161. [DOI] [PubMed] [Google Scholar]
- 2.Stetler-Stevenson W G, Aznavoorian S, Liotta L A. Annu Rev Cell Biol. 1993;9:541–573. doi: 10.1146/annurev.cb.09.110193.002545. [DOI] [PubMed] [Google Scholar]
- 3.Chen W-T, Lee C C, Goldstein L, Bernier S, Liu C H, Lin C Y, Yeh Y, Monsky W L, Kelly T, Dai M, Mueller S C. Breast Cancer Res Treat. 1994;31:217–226. doi: 10.1007/BF00666155. [DOI] [PubMed] [Google Scholar]
- 4.Cockett M I, Birch M L, Murphy G, Hart I R, Docherty A J P. Biochem Soc Trans. 1994;22:55–57. doi: 10.1042/bst0220055. [DOI] [PubMed] [Google Scholar]
- 5.Birkedal-Hansen H, Moore W G I, Bodden M K, Windsor L J, Birkedal-Hansen B, DeCarlo A, Engler J A. Crit Rev Oral Biol Med. 1993;4:197–250. doi: 10.1177/10454411930040020401. [DOI] [PubMed] [Google Scholar]
- 6.Azzam H S, Thompson E W. Cancer Res. 1992;52:4540–4544. [PubMed] [Google Scholar]
- 7.Monsky W L, Kelly T, Lin C Y, Yeh Y, Stetler-Stevenson W G, Mueller S C, Chen W-T. Cancer Res. 1993;53:3159–3164. [PubMed] [Google Scholar]
- 8.Yu M, Sato H, Seiki M, Thompson E W. Cancer Res. 1995;55:3272–3277. [PubMed] [Google Scholar]
- 9.Azzam H S, Arand G, Lippman M E, Thompson E W. J Natl Cancer Inst. 1993;85:1758–1764. doi: 10.1093/jnci/85.21.1758. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Fu X, Brown P D, Crimmin M J, Hoffman R M. Cancer Res. 1994;54:4726–4728. [PubMed] [Google Scholar]
- 11.Horton R M, Hunt H D, Ho S N, Pullen J K, Pease L R. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 12.Strongin A Y, Collier I, Bannikov G, Marmer B L, Grant G A, Goldberg G I. J Biol Chem. 1995;270:5331–5338. doi: 10.1074/jbc.270.10.5331. [DOI] [PubMed] [Google Scholar]
- 13.Sato H, Takino T, Okada Y, Cao J, Shinagawa A, Yamamoto E, Seiki M. Nature (London) 1994;370:61–65. doi: 10.1038/370061a0. [DOI] [PubMed] [Google Scholar]
- 14.Okada A, Bellocq J P, Rouyer N, Chenard M P, Rio M C, Chambon P, Basset P. Proc Natl Acad Sci USA. 1995;92:2730–2734. doi: 10.1073/pnas.92.7.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato H, Takino T, Kinoshita T, Seiki M. FEBS Lett. 1996;385:238–240. doi: 10.1016/0014-5793(96)00389-4. [DOI] [PubMed] [Google Scholar]
- 16.Pei D, Weiss S J. J Biol Chem. 1996;271:9135–9140. doi: 10.1074/jbc.271.15.9135. [DOI] [PubMed] [Google Scholar]
- 17.Imai K, Ohuchi E, Aoki T, Nomura H, Fujii Y, Sato H, Seiki M, Okada Y. Cancer Res. 1996;56:2707–2710. [PubMed] [Google Scholar]
- 18.Ohuchi E, Imai K, Fujii Y, Sato H, Seiki M, Okada Y. J Biol Chem. 1997;272:2446–2451. doi: 10.1074/jbc.272.4.2446. [DOI] [PubMed] [Google Scholar]
- 19.Monsky W L, Lin C-Y, Aoyama A, Kelly T, Mueller S C, Akiyama S K, Chen W-T. Cancer Res. 1994;54:5702–5710. [PubMed] [Google Scholar]
- 20.Morphy J R, Millican T A. Curr Med Chem. 1996;2:743–762. [Google Scholar]
- 21.Aoyama A, Chen W-T. Proc Natl Acad Sci USA. 1990;87:8296–8300. doi: 10.1073/pnas.87.21.8296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen W-T, Yeh Y, Nakahara H. J Tissue Cult Method. 1994;16:177–181. [Google Scholar]
- 23.Cao J, Sato H, Takino T, Seiki M. J Biol Chem. 1995;270:801–805. doi: 10.1074/jbc.270.2.801. [DOI] [PubMed] [Google Scholar]
- 24.Mueller S C, Yeh Y, Chen W-T. J Cell Biol. 1992;119:1309–1325. doi: 10.1083/jcb.119.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen W-T. J Exp Zool. 1989;251:167–185. doi: 10.1002/jez.1402510206. [DOI] [PubMed] [Google Scholar]
- 26.Chen W-T. Enzyme Protein. 1996;49:59–71. doi: 10.1159/000468616. [DOI] [PubMed] [Google Scholar]
- 27.Nakahara H, Nomizu M, Akiyama S K, Yamada Y, Yeh Y Y, Chen W T. J Biol Chem. 1996;271:27221–27224. doi: 10.1074/jbc.271.44.27221. [DOI] [PubMed] [Google Scholar]
- 28.Pineiro-Sanchez M L, Goldstein L A, Dodt J, Howard L, Yeh Y, Chen W. J Biol Chem. 1997;272:7595–7601. doi: 10.1074/jbc.272.12.7595. [DOI] [PubMed] [Google Scholar]
- 29.Tryggvason K, Hoyhtya M, Pyke C. Breast Cancer Res Treat. 1993;24:209–218. doi: 10.1007/BF01833261. [DOI] [PubMed] [Google Scholar]
- 30.Steffensen B, Wallon U M, Overall C M. J Biol Chem. 1995;270:11555–11566. doi: 10.1074/jbc.270.19.11555. [DOI] [PubMed] [Google Scholar]
- 31.Mackay A R, Gomez D E, Cottam D W, Rees R C, Nason A M, Thorgeirsson U P. BioTechniques. 1993;15:1048–1051. [PubMed] [Google Scholar]