Abstract
The spindle checkpoint monitors mitotic spindle integrity and the attachment of kinetochores to the spindle. Upon sensing a defect the checkpoint blocks cell cycle progression and thereby prevents chromosome missegregation. Previous studies in budding yeast show that the activated spindle checkpoint inhibits the onset of anaphase by an unknown mechanism. One possible target of the spindle checkpoint is anaphase promoting complex (APC), which controls all postmetaphase events that are blocked by spindle checkpoint activation. We have isolated mad2, a spindle checkpoint component in fission yeast, and shown that mad2 overexpression activates the checkpoint and causes a cell cycle arrest at the metaphase-to-anaphase transition. In addition to the observation that mad2-induced arrest can be partially relieved by mitosis-promoting factor inactivation, we present genetic evidence consistent with the hypothesis that the spindle checkpoint imposes a cell cycle arrest by inhibiting APC-dependent proteolysis.
The precision of the cell division process in eukaryotic organisms is enhanced by checkpoints that prevent the initiation of critical steps in the cell cycle until prior steps have been accurately completed (1). In the absence of functional checkpoints, DNA damage and spindle defects would escape detection, leading to the accumulation of errors in key cell cycle events such as DNA replication and chromosome segregation (2).
The spindle-assembly checkpoint ensures accurate nuclear division by monitoring the integrity of the mitotic spindle and the proper attachment of chromosomes to the spindle, and then preventing the onset of anaphase in the presence of a defective mitotic spindle or unattached kinetochores (3–6). Characterization of the spindle-assembly checkpoint began with the isolation of mutants in the budding yeast Saccharomyces cerevisiae, in which the presence of spindle damage fails to arrest cell cycle progression (7, 8). Eight spindle checkpoint genes have been identified in budding yeast, MAD1–3, BUB1–3, MPS1, and CDC55 (3). Homologues of several checkpoint components subsequently have been identified in other species, suggesting that the spindle checkpoint system is evolutionarily conserved (3). Activation of the spindle checkpoint in budding yeast blocks cells with condensed chromosomes prior to the onset of anaphase (7, 9, 10). However, because cell cycle stages between S phase and anaphase are not morphologically distinct in budding yeast (11), the precise point at which the spindle checkpoint arrests cell cycle progression is not well defined cytologically. Also unknown are the mechanism by which the checkpoint is activated and the mechanism by which activation of this pathway blocks cell cycle progression.
Considerable evidence suggests that the onset of anaphase (sister chromatid separation) and other postmetaphase events (spindle elongation and exit from mitosis) are controlled by proteolysis of specific proteins by the ubiquitination pathway (6–9). Sister chromatid separation requires the degradation of Pds1p in budding yeast (12) and Cut2p in fission yeast (13). Telophase progression and mitotic exit require the degradation of mitotic cyclin B (14). The only element in this ubiquitination pathway whose activity is cell-cycle regulated is the ubiquitin ligase (E3), also called the cyclosome or anaphase promoting complex (APC) (15–17). APC catalyzes the transfer of ubiquitin from ubiquitin-conjugating enzymes to the substrate proteins (15), which are subsequently degraded by the 26S proteasome (18). APC, a 20S protein complex in Xenopus, contains eight protein subunits (19). Four of the subunits have also been identified in S. cerevisiae: CDC16, CDC23, CDC27, and APC1 (19). Fission yeast cut9, nuc2, and cut4 (20) encode homologues of budding yeast Cdc16p, Cdc27p, and Apc1p, respectively, and are also required for anaphase initiation (19). Because no postmetaphase events take place when the spindle checkpoint is activated, it is possible that APC-dependent proteolysis is prevented by the checkpoint either by inhibiting the ubiquitination of the target proteins or by blocking the access of substrates to the proteolysis machinery (4).
In a genetic screen in the fission yeast Schizosaccharomyces pombe designed to identify cDNAs that cause mitotic defects when overproduced (X.H., T.E.P., and S.S., unpublished data), we have isolated a cDNA encoding the fission yeast homologue of the budding yeast spindle checkpoint protein Mad2p. We show here that fission yeast mad2p is required for spindle checkpoint function and that overexpression of mad2 in fission yeast activates the spindle checkpoint. This activation causes a cell cycle arrest at the metaphase-to-anaphase transition, as judged by both morphological and biochemical criteria. Consistent with the hypothesis that activation of the spindle checkpoint pathway arrests the cell cycle at metaphase by inhibiting APC-dependent protein degradation, we demonstrate a genetic interaction between APC and mad2 and show that the mad2-induced cell cycle arrest is partially relieved by inactivation of mitosis-promoting factor (MPF) kinase.
MATERIALS AND METHODS
Yeast Strains and Genetic Analysis.
S. pombe strains used were the haploid wild-type strain (h−, leu1–32, ura4-D18, ade6-m216) (21), diploid wild-type strain (h−/h+, leu1–32/leu1–32, ura4-D18/ura4-D18, ade6-m210/ade6-m216) (21), mutant strains nuc2ts(h− nuc2–662, leu1–32, ura4-D18, ade6-m216) (22), cut9ts(h− cut9–665, leu1–32) (23), nda3cs(h− nda3–311, leu1–32, ura4-D18, ade6-m216) (24), and cdc2–33ts(h-cdc2–33ts, leu1–32, ura4-D18) (25). The mad2 deletion strains (h− or h+, mad2::ura4, leu1–32, ade6-m210) were generated by replacing the restriction fragment SpeI/HindIII, which corresponds to amino acids 90–203, in one copy of the mad2 gene with the ura4 gene in a wild-type diploid, and were confirmed by Southern blot analysis. Tetrad analysis showed cosegregation of ura+ with thiabendazole (TBZ) (Sigma) hypersensitivity at thiabendazole concentrations of 20–50 μg/ml. The mad2Δ nda3cs double mutant was generated by crossing mad2Δ with nda3–311cs and was identified by its cold-sensitive(cs) and ura+ phenotypes. Yeast culture, transformation, and genetic manipulations were performed by standard procedures (26).
DNA Methods.
In a screen described in detail elsewhere (X.H., T.E.P., and S.S., unpublished data), a S. pombe cDNA library in pREP3X vector (a gift from Bruce Edgar, Fred Hutchinson Cancer Research Center, and Chris Norbury, University of Oxford, U.K.), in which cDNA expression is controlled by the thiamine-repressible nmt1 promoter (27), was transformed into wild-type cells. Transformants were screened based on the toxicity and the mitotic defects caused by cDNA overexpression. Plasmids from S. pombe transformants were recovered by standard procedures (26) and amplified in Escherichia coli. The cDNA inserts and their restriction fragments were subcloned into Bluescript KS(−) (Stratagene) and sequenced using the Sequenase Version 2.0 (United States Biochemical). pREP41X-mad2 and pREP81X-mad2 were constructed by subcloning the XhoI/BamHI fragment of the mad2 cDNA from pREP3X into pREP41X and pREP81X, respectively, to achieve lower levels of mad2 overexpression (27).
H1 Kinase Assay.
Wild-type cells transformed with pREP3X-mad2 (mad2 OP) were transferred from medium with thiamine, in which transcription is repressed, to thiamine-free medium for 13 hr to induce mad2 overexpression at 32°C. The wild-type control cells that were transformed with the vector pREP3X were treated identically. Total yeast extracts were prepared using the glass-bead method (26). Equal amounts of total protein extract (determined by Bradford assay, Bio-Rad) from mad2 overexpressing and control cells were subjected to immunoprecipitation using anti-cdc2 antiserum 4711 (a gift from Kathy Gould, Vanderbilt University, Nashville, TN), following previously described procedures (28). One-half of each immunoprecipitated product was subjected to an H1 kinase assay (29); the other half was used for Western blot analysis (ECL, Amersham) to determine the amount of precipitated p34cdc2.
Cell Viability Test.
Equal numbers of single mutant cells, mad2Δ or nda3cs, and double mutant cells, mad2Δ nda3cs, were spread on YE plates. The plates were then incubated at the nda3cs-restrictive temperature of 18°C for 0, 3, 6, and 9 hr and then returned to the permissive temperature of 32°C. The number of colonies was counted after 3 days.
DNA Content Analysis.
mad2Δ or nda3cs and double mutant cells mad2Δ nda3cs cells were grown in YE liquid medium at 32°C to early log phase and shifted to 20°C. At each time point, cell samples were fixed with ethanol, treated with RNase, and stained with propidium iodide, and their DNA content was measured by flow cytometry (30).
Fluorescence Microscopy.
Immunofluorescence procedures were as described (31). Cells were fixed with glutaraldehyde and paraformaldehyde and stained with anti-tubulin monoclonal antibody TAT1 (32) (a gift from Keith Gull, University of Manchester Institute of Science and Technology, U.K.) and fluorescein-conjugated anti-mouse secondary antibody (Pierce) to reveal the microtubule structures. Cells were simultaneously stained with 4′,6-diamidino-2-phenylindole (DAPI) to visualize the DNA.
RESULTS
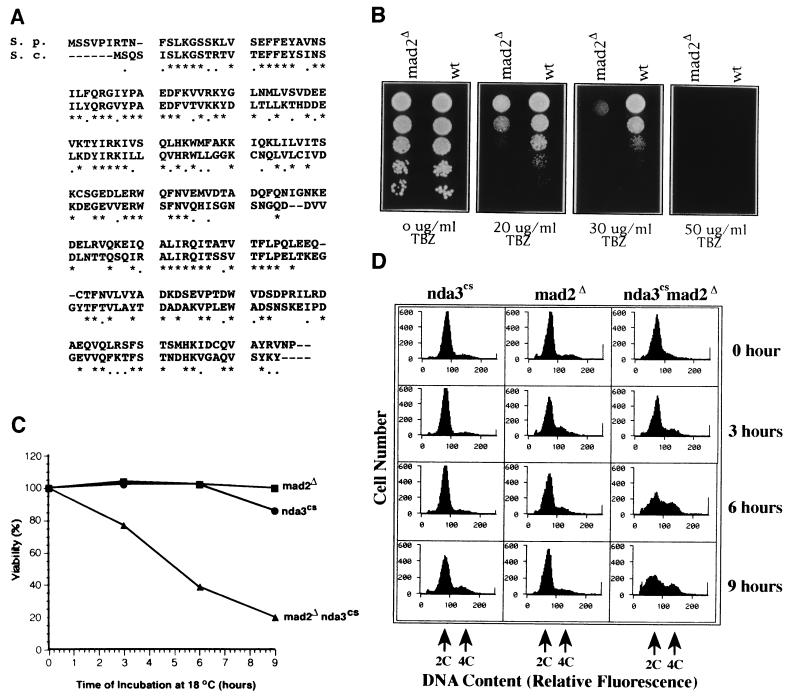
In a cDNA library overproduction screen in which cells displaying transcription-dependent lethality were examined microscopically, mad2 was isolated as a gene whose overproduction arrests cells in mitosis (X.H., T.E.P., and S.S., unpublished data). The amino acid sequence of fission yeast mad2p is 53% identical to budding yeast Mad2p (GenBank accession no. U14132) (Fig. 1A). If mad2p functions as a spindle checkpoint protein in fission yeast, cells lacking mad2p would be expected to be hypersensitive to microtubule-destabilizing drugs, such as TBZ, because of their inability to arrest cell cycle progression in the presence of a defective spindle (7, 8). A haploid strain containing a mad2 deletion mutation (mad2Δ) was viable, but the strain was hypersensitive to TBZ (Fig. 1B).
Figure 1.
S. pombe mad2p functions as a spindle-assembly checkpoint protein. (A) Open reading frame of S. pombe mad2 (GenBank accession no. U72150) aligned with the S. cerevisiae Mad2p (GenBank accession no. U14132). Identical amino acids are labeled with stars and conserved amino acids are labeled with dots. (B) Hypersensitivity of mad2Δ to thiabendazole (TBZ) treatment. Serial dilutions (1/5) of an equal number of mad2Δ and wild-type cells were spotted on Edinburgh minimum medium plates with the indicated concentration of TBZ. (C) Viability test of the single mutants mad2Δ and nda3–311cs and the double mutant mad2Δ nda3–311cs after a transient incubation at 18°C. Colonies on each plate were counted and the percentage of viable cells was calculated by normalization to the colony number of the 0-hr samples. (D) Flow-cytometric analysis of DNA content of the single mutants mad2Δ and nda3–311cs and the double mutant mad2Δ nda3–311cs after a transient incubation at 20°C. The relative fluorescence corresponding to 2C and 4C DNA content is marked by arrows; hours indicate the length of 20°C incubation.
To determine whether mad2Δ was also sensitive to spindle defects of a different kind, we tested the synthetic lethality of mad2Δ and nda3–311cs. nda3–311cs mutants at the restrictive temperature are arrested at prometaphase and lack a mitotic spindle due to a cold-sensitive mutation in the β-tubulin gene. The nda3–311cs phenotype is highly reversible: within 6 min after return to the permissive temperature, the cells form a functional mitotic spindle and complete mitosis properly (33). Both mad2Δ and nda3–311cs, as single mutants, maintained high viability with cold treatment. In contrast, the mad2Δnda3–311cs double mutant rapidly lost viability (Fig. 1C). Under these conditions the double mutant, but neither of the single mutants, continued to synthesize DNA, as evidenced by the appearance of cells with a greater than 2C DNA content (Fig. 1D). Microscopic observation of the cells stained with DAPI showed that after 9 hr at the restrictive temperature, 33% of the double mutant cells had abnormally large nuclei and very bright DNA fluorescence. In 59% of cells, septation proceeded in the absence of nuclear division, resulting in the “cut” phenotype (34) and an unequal partitioning of DNA to the daughter cells. These microscopic images are consistent with the histograms of DNA content (Fig. 1D) and provide an explanation for the origin of heteroploidy in the double mutant. mad2Δ cells, therefore, bypass mitosis in the absence of a normal spindle and undergo both septation and the next round of DNA replication.
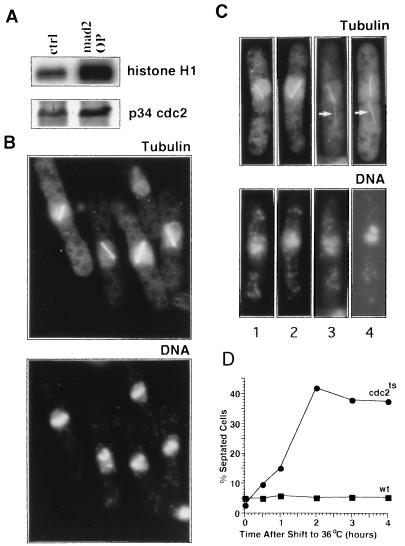
Overexpression of the mad2 cDNA in fission yeast, under control of the thiamine-repressible promoter nmt1 (35), inhibited cell growth. Thirteen hours after transcriptional derepression of mad2 at 32°C, cell division ceased and the arrested cells displayed characteristic features of metaphase: the H1 kinase activity of MPF was significantly higher than that of wild-type asynchronous cells when normalized to the amount of immunoprecipitated p34cdc2 (Fig. 2A), and >50% of the cells had condensed chromosomes and a short mitotic spindle (Fig. 2B), compared with only 2% in a wild-type asynchronous culture (31).
Figure 2.
mad2 overexpression arrests cells at the metaphase-to-anaphase transition, and subsequent cdc2ts inactivation promotes septum formation. (A) MPF H1 kinase assay. MPF protein complex was immunoprecipitated from total protein extract by anti-cdc2 antiserum and subjected to H1 kinase assay. (Upper) Autoradiography of [γ-32P]ATP-labeled histone H1. (Lower) Western blot of immunoprecipitated p34 cdc2. (B Upper) Wild-type cells overexpressing mad2 in which microtubules were visualized with anti-tubulin indirect immunofluorescence. (Lower) The same cells stained with DAPI to visualize DNA. (C) cdc2–33ts and wild-type cells, arrested by mad2 overexpression at 25°C, were shifted to 36°C to inactivate MPF. Tubulin (Upper) and DNA (Lower) were visualized as in B. Arrows in cells 3 and 4 (Upper) indicate cytoplasmic microtubules. (D) cdc2–33ts and wild-type cells, arrested by mad2 overexpression at 25°C, were shifted to 36°C to inactivate MPF. Cells were fixed with ethanol, and the percentage of septated cells was counted using light microscopy.
The high MPF activity in cells overexpressing mad2 could be either a cause or a consequence of the metaphase arrest resulting from mad2 overexpression. To distinguish between these possibilities, we investigated the effects of MPF inactivation on cell cycle arrest induced by mad2 overexpression. The mad2 cDNA was transformed into cdc2–33ts cells, which contain a mutation in the MPF kinase catalytic subunit that results in rapid, temperature-sensitive loss of kinase activity (28, 36) and cell cycle arrest after 30 min at the restrictive temperature (37). In both wild-type cells (Fig. 2B) and the cdc2–33ts mutant grown at the permissive temperature (data not shown), mad2 overexpression arrested cells at metaphase with a linear array of brightly stained condensed chromosomes and a short mitotic spindle. The cells were then transferred to the restrictive temperature to inactivate MPF. After 1 hr, the chromosomes remained condensed and unseparated and the mitotic spindle remained intact and failed to elongate. The most striking abnormality was the relocalization of the undivided nucleus to one-half of the cell (Fig. 2C), whereas in wild-type cells, relocalization occurs only after nuclear division. In some of these cells (Fig. 2C, cells 3 and 4), cytoplasmic microtubule structures started to form (Fig. 2C, arrows in cells 3 and 4) before the short spindle completely disassembled, although in wild-type cells, cytoplasmic microtubule reassembly occurs only after mitotic spindle elongation and disassembly (31). After 2 hr, 40–45% of the arrested cells had a medial septum (Fig. 2D), one anucleated cell, and one uninucleated cell with decondensed chromosomes. Anti-tubulin immunofluorescence revealed that these septated cells no longer had a mitotic spindle but contained cytoplasmic microtubules (data not shown). MPF inactivation in cdc13–117ts, a strain harboring a mutation in the cyclin B subunit of MPF (38), had similar effects on mad2 arrested cells (data not shown).
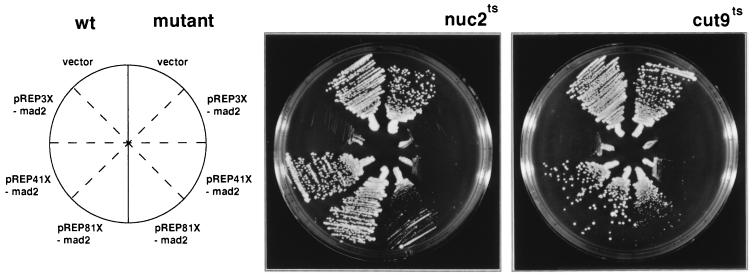
mad2 overexpression prevented chromosome separation, spindle elongation, and exit from mitosis, suggesting that it blocked all aspects of the metaphase-to-anaphase transition. APC, a protein complex that functions as a ubiquitin ligase in the proteolysis pathway, controls the metaphase-to-anaphase transition. Fission yeast cells with a temperature-sensitive mutation in the APC subunits cut4, cut9, or nuc2 transiently arrest in metaphase at the restrictive temperature with phenotypes that resemble the metaphase arrest induced by mad2 overexpression in wild-type cells (20, 22, 23). These mutant cells eventually exit from mitosis without undergoing anaphase (20, 22, 23), and their terminal phenotypes are similar to mad2-arrested cells in which MPF has subsequently been inactivated: the cells are septated, have a single nucleus in one of the daughter cells, and do not undergo spindle elongation. Because mad2 overexpression blocks the metaphase-to-anaphase transition, it is possible that activation of the spindle checkpoint arrests cell cycle progression by inhibiting APC activity. This hypothesis predicts a genetic interaction between the spindle checkpoint and protein ubiquitination systems. To test this possibility we asked whether mutants in APC are hypersensitive to mad2 overexpression when compared with wild-type cells (Fig. 3). Medium-level expression of mad2 (pREP41X-mad2) had no obvious effect on wild-type cell growth but almost completely inhibited the growth of both cut9–665ts and nuc2–663ts cells at the permissive temperature (25°C). Low-level expression (pREP81X-mad2) also specifically inhibited the mutants. As expected, high-level expression of mad2 (pREP3X-mad2) was lethal to all three strains.
Figure 3.
APC mutants are hypersensitive to mad2 overexpression. Wild-type cells and APC subunit mutants (cut9–665ts and nuc2–663ts) were transformed with pREP3X vector and a set of mad2 overexpression constructs: pREP3X-mad2, pREP41X-mad2, and pREP81X-mad2. Transformants were streaked on EMM plates in the absence of thiamine so that three levels of mad2 overexpression were achieved via the three different strength nmt1 promoters: pREP3X-mad2, high; pREP41X-mad2, medium; and pREP81X-mad2, low (27). On each plate, wild-type transformants were streaked on the left and the mutant transformants on the right. The plates were incubated at 25°C.
DISCUSSION
mad2p Functions as a Spindle Checkpoint Protein in Fission Yeast.
We have identified mad2 in fission yeast and shown that it encodes a protein with high sequence similarity to the Mad2p spindle checkpoint protein from budding yeast (GenBank accession no. U14132), frog (39), and human (6). Like Mad2p in budding yeast, fission yeast mad2p is not essential for cell division and functions as a spindle checkpoint protein, because the mad2 deletion is viable but hypersensitive to the microtubule-destabilizing drug thiabendazole. More importantly, nda3cs cells in which mad2 is deleted fail to arrest at metaphase although they lack a functional mitotic spindle at the restrictive temperature. This indicates that fission yeast mad2p functions as a spindle checkpoint protein that is essential for cell cycle arrest in response to a defective spindle.
mad2 Overexpression Causes a Cell Cycle Block at the Metaphase-to-Anaphase Transition.
In wild-type fission yeast cells, mad2 overexpression causes a cell cycle arrest at a metaphase like stage: cells have hypercondensed chromosomes and a short mitotic spindle; the nucleus is undivided; and the MPF kinase activity is high. Except for the absence of a mitotic spindle in the β-tubulin mutant, this metaphase arrest resembles the cell cycle block in nda3cs mutants (33) and cells treated with microtubule destabilizing drugs (40), indicating that mad2 overexpression mimics normal checkpoint activation by a defective spindle. Furthermore, because mad2 overexpression can arrest cell cycle progression in the presence of an apparently normal spindle structure (Fig. 2B), we were able to precisely determine that activation of the spindle checkpoint in fission yeast arrests cell cycle progression at the metaphase-to-anaphase transition. Subsequent MPF inactivation after the mad2-induced arrest causes the cells to bypass anaphase and directly exit from mitosis (see Fig. 4 for a model) apparently without either spindle elongation or sister chromatid separation. This latter conclusion is based on the absence of nuclear division as monitored by DAPI staining of the DNA but has not been verified using fluorescense in situ hybridization analysis (41). These results lend further support to the conclusion that the mad2-induced arrest is at the metaphase-to-anaphase transition and is consistent with the proposal that anaphase and exit from mitosis are independently regulated (17). Previously, it was known that mitotic cyclin B degradation, thus the inactivation of MPF, is necessary for telophase progression (14). Our results suggest that MPF inactivation is both necessary and sufficient to promote both telophase and cytokinesis. Consistent with our results, budding yeast cells arrested by nocodazole, a drug that destabilizes microtubules and thereby activates the spindle checkpoint, can bypass anaphase and complete the next S phase when MPF is inactivated (42).
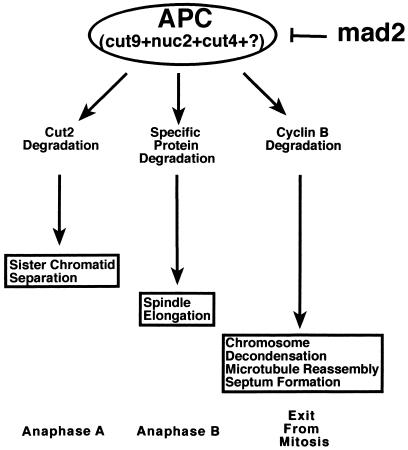
Figure 4.
A model of the mechanism by which mad2 overexpression blocks the metaphase-to-anaphase transition. APC targets specific proteins for degradation, which is required for three different postmetaphase events. Cyclin B degradation is required for the exit from mitosis. Cut2p degradation is required for sister chromosome separation. The protein whose degradation is required for spindle elongation has not yet been identified. mad2 overexpression mimics activation of the spindle checkpoint and blocks the metaphase-to-anaphase transition by directly or indirectly inhibiting APC.
The Spindle Checkpoint May Inhibit APC-Dependent Proteolysis.
It was previously proposed that APC, which biochemically functions as a ubiquitin ligase, controls several aspects of the metaphase-to-anaphase transition by targeting specific proteins for degradation (17) (Fig. 4). Substrates of APC include cyclin B, whose degradation is required for MPF inactivation and the exit from mitosis in budding yeast and frog eggs (15, 17); Cut2p, whose degradation is required for sister chromatid separation in fission yeast (13); and a yet to be identified protein(s), whose degradation is required for spindle elongation. Pds1p, an anaphase-inhibitor protein in S. cerevisiae, is also a target of APC (43). It has been hypothesized that the spindle-assembly checkpoint may prevent the onset of anaphase by interfering with specific protein degradation (4). We have demonstrated that two strains, which have mutations in different APC subunits, are hypersensitive to mad2 overproduction. These genetic results are consistent with the model that spindle checkpoint activation inhibits APC activity (Fig. 4). Furthermore, in nocodazole-treated budding yeast cells, Pds1p-dependent sister chromatid separation (12) and B-type cyclin ubiquitination activity (44) are inhibited. In frog egg extracts, activation of the spindle checkpoint pathway also stabilizes cyclin B and maintains high MPF H1 kinase activity (45). Consistent with these observations, we have also detected high MPF activity following mad2 overexpression, indicating that cyclin B degradation is inhibited. Based on these results and the genetic interaction between mad2 and components of the APC, we propose that activation of the mad2-dependent spindle checkpoint pathway in fission yeast blocks the metaphase-to-anaphase transition by inhibiting APC-dependent proteolysis (Fig. 4). However, we cannot distinguish between inhibition of ubiquitination activity and protection of proteins from the proteolytic machinery. We anticipate that future studies, especially those using in vitro biochemical approaches, will elucidate the precise biochemical mechanism of this inhibition.
Our model also explains the previous unexpected observations that cdc13ts mutants are hypersensitive (46) and the 26S proteasome mutants, mts2ts and mts3ts, are resistant (47) to microtubule-destabilizing drug treatment. Because cdc13ts cells fail to maintain high MPF activity (36), they may not be properly arrested by spindle checkpoint activation, which would result in lethality. Activation of the spindle checkpoint arrests cells at metaphase by inhibiting APC and thereby interfering with specific protein degradation. Because untreated mts mutants already have reduced ubiquitin-mediated proteolysis activity (47, 48), these cells would be expected to have an enhanced response to the spindle damage caused by TBZ treatment.
In budding yeast, eight genes (MAD1–3, BUB1–3, MPS1, and CDC55) have been identified as components of the spindle checkpoint pathway (3). It was shown recently that Mad2p in frog and human localizes to unattached kinetochores, indicating that it may detect the defects in the attachment of kinetochores to the spindle (6, 39). Furthermore, MAD1, MAD3, and BUB2 function downstream of MAD2 in the spindle checkpoint pathway (49), indicating, by analogy, that fission yeast mad2p may not directly interact with APC. Overexpression of mad2 in fission yeast may inhibit APC by activating downstream genes in the spindle checkpoint pathway. It is therefore possible that overexpression of other spindle checkpoint genes would result in phenotypes similar to those caused by mad2 overexpression. In fact, while this manuscript was in preparation, it was reported that overexpression of MPS1 in wild-type budding yeast cells activates the spindle checkpoint pathway (9). In the screen that led to the identification of mad2 (X.H., et al., unpublished data), we also identified and are currently characterizing two other new fission yeast genes with overproduction phenotypes that mimic that of mad2 overexpression. One of these cDNAs encodes a protein with a 269-aa region with 45% sequence identity to the kinase domain of the dual-specificity protein kinase family (50–52), which includes the budding yeast Mps1p. The similarity between the overexpression phenotypes of mad2 and these two new genes makes them excellent candidates for additional components of the spindle checkpoint system in fission yeast.
Acknowledgments
We are grateful to Ngoctuyen Ong for technical assistance, Keith Gull for providing the TAT 1 antibody, Kathy Gould for the 4711 antibody, Chris Norbury and Bruce Edgar for the cDNA library, and Iain Hagan, Dick McIntosh, Steve Elledge, Ursula Fleig, and Kathy Gould for insightful discussions and comments on the manuscript. This work was supported by a grant from the National Institutes of Health to S.S. (GM49119). T.E.P. was supported by a National Institute of Aging Training Grant (AG00183).
ABBREVIATIONS
- APC
anaphase-promoting complex
- MPF
mitosis-promoting factor
- TBZ
thiabendazole
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U72150).
References
- 1.Harwell L H, Weinert T A. Science. 1989;266:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 2.Paulovich A G, Toczyski D P, Hartwell L H. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 3.Rudner A D, Murray A W. Curr Opin Cell Biol. 1996;8:773–780. doi: 10.1016/s0955-0674(96)80077-9. [DOI] [PubMed] [Google Scholar]
- 4.Wells W A E. Trends Cell Biol. 1996;6:228–234. doi: 10.1016/0962-8924(96)10018-0. [DOI] [PubMed] [Google Scholar]
- 5.Chen R, Waters J C, Salmon E D, Murray A W. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, Benezra R. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- 7.Li R, Murray A W. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 8.Hoyt M A, Tolis L, Roberts B T. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 9.Hardwick K G, Weiss E, Luca F C, Winey M, Murray A W. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- 10.Guacci V, Hogan E, Koshland D. J Cell Biol. 1994;125:517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winey M, Mamay C L, O’Toole E T, Mastronarde D N, Giddings T H, Jr, McDonald K L, McIntosh J R. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamamoto A, Guacci V, Koshland D. J Cell Biol. 1996;133:99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Funabiki H, Yamano H, Kumada K, Nagao K, Hunt T, Yanagida M. Nature (London) 1996;381:438–441. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- 14.Holloway S L, Glotzer M, King R W, Murray A W. Cell. 1993;73:1393–1402. doi: 10.1016/0092-8674(93)90364-v. [DOI] [PubMed] [Google Scholar]
- 15.King R W, Peters J M, Tugendreich S, Rolfe M, Hieter P, Kirschner M W. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- 16.Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca F C, Ruderman J V, Hershko A. Mol Biol Cell. 1995;6:185–197. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irniger S, Piatti S, Michaelis C, Nasmyth K. Cell. 1995;81:269–278. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- 18.Peters J-M. Trends Biochem Sci. 1994;9:377–382. doi: 10.1016/0968-0004(94)90115-5. [DOI] [PubMed] [Google Scholar]
- 19.King R W, Deshaies R J, Peters J-M, Kirschner M W. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 20.Yamashita Y M, Nakaseko Y, Samejima I, Kumada K, Yamada H, Michaelson D, Yanagida M. Nature (London) 1996;384:276–279. doi: 10.1038/384276a0. [DOI] [PubMed] [Google Scholar]
- 21.Minshull J, Straight A, Rudner A D, Dernburg A F, Belmon A, Murray A W. Curr Biol. 1996;6:1609–1620. doi: 10.1016/s0960-9822(02)70784-7. [DOI] [PubMed] [Google Scholar]
- 22.Hirano T, Hiraoka Y, Yanagida M. J Cell Biol. 1988;106:1171–1183. doi: 10.1083/jcb.106.4.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samejima I, Yanagida M. J Cell Biol. 1994;127:1655–1670. doi: 10.1083/jcb.127.6.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hiraoka Y, Toda T, Yanagida M. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- 25.Carr A M, MacNeill S A, Hayles J, Nurse P. Mol Gen Genet. 1989;218:41–49. doi: 10.1007/BF00330563. [DOI] [PubMed] [Google Scholar]
- 26.Moreno S, Klar A, Nurse P. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 27.Forsburg S L. Nucleic Acids Res. 1993;21:2955–2956. doi: 10.1093/nar/21.12.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moreno S, Hayles J, Nurse P. Cell. 1989;58:361–372. doi: 10.1016/0092-8674(89)90850-7. [DOI] [PubMed] [Google Scholar]
- 29.Conell-Crowley L, Solomon M J, Wei N, Harper J W. Mol Biol Cell. 1993;4:79–92. doi: 10.1091/mbc.4.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sazer S, Sherwood S. J Cell Sci. 1990;97:509–516. doi: 10.1242/jcs.97.3.509. [DOI] [PubMed] [Google Scholar]
- 31.Hagan I, Hyams J. J Cell Sci. 1988;89:343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- 32.Woods A, Sherwin T, Sasse R, Macrae T H, Baines A J, Gull K. J Cell Sci. 1989;93:491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]
- 33.Kanbe T, Hiraoka Y, Tanaka K, Yanagida M. J Cell Sci. 1990;96:275–182. doi: 10.1242/jcs.96.2.275. [DOI] [PubMed] [Google Scholar]
- 34.Hirano T, Funahashi S, Uemura T, Yanagida M. EMBO J. 1986;5:2973–2979. doi: 10.1002/j.1460-2075.1986.tb04594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maundrell K. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- 36.Booher R N, Alfa C E, Hyams J S, Beach D H. Cell. 1989;57:485–497. doi: 10.1016/0092-8674(89)90429-7. [DOI] [PubMed] [Google Scholar]
- 37.Nurse P, Thuriaux P, Nasmyth K. Mol Gen Genet. 1976;146:167–178. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- 38.Hagan I, Hayles J, Nurse P. J Cell Sci. 1988;91:587–595. doi: 10.1242/jcs.91.4.587. [DOI] [PubMed] [Google Scholar]
- 39.Chen R-H, Waters J C, Salmon E D, Murray A W. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- 40.Walker G M. J Gen Microbiol. 1982;128:61–71. doi: 10.1099/00221287-128-1-61. [DOI] [PubMed] [Google Scholar]
- 41.Uzawa S, Yanagida M. J Cell Sci. 1992;101:267–275. doi: 10.1242/jcs.101.2.267. [DOI] [PubMed] [Google Scholar]
- 42.Dahmann C, Diffley J F X, Nasmyth K. Curr Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- 43.Funabiki H, Hagan I, Uzawa S, Yanagida M. J Cell Biol. 1993;121:961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zachariae W, Nasmyth K. Mol Biol Cell. 1996;7:791–801. doi: 10.1091/mbc.7.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minshull J, Sun H, Tonks N K, Murray A W. Cell. 1994;79:475–486. doi: 10.1016/0092-8674(94)90256-9. [DOI] [PubMed] [Google Scholar]
- 46.Booher R, Beach D. EMBO J. 1988;7:2321–2327. doi: 10.1002/j.1460-2075.1988.tb03075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gordon C, McGurk G, Wallace M, Hastie N D. J Biol Chem. 1996;271:5704–5711. doi: 10.1074/jbc.271.10.5704. [DOI] [PubMed] [Google Scholar]
- 48.Gordon C, McGurk G, Dillon P, Rosen C, Hastie N D. Nature (London) 1993;366:355–357. doi: 10.1038/366355a0. [DOI] [PubMed] [Google Scholar]
- 49.Hardwick K G, Murray A W. J Cell Biol. 1995;131:709–720. doi: 10.1083/jcb.131.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mills G B, Schmandt R, McGill M, Amendola A, Hill M, Jacobs K, May C, Rodricks A M, Campbell S, Hogg D. J Biol Chem. 1992;267:16000–16006. [PubMed] [Google Scholar]
- 51.Douville E M, Afar D E, Howell B W, Letwin K, Tannock L, Ben-David Y, Pawson T, Bell J C. Mol Cell Biol. 1992;12:2681–2689. doi: 10.1128/mcb.12.6.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lauze E, Stoelcker B, Luca F C, Weiss E, Schutz A R, Winey M. EMBO J. 1995;14:1655–1663. doi: 10.1002/j.1460-2075.1995.tb07154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]