Abstract
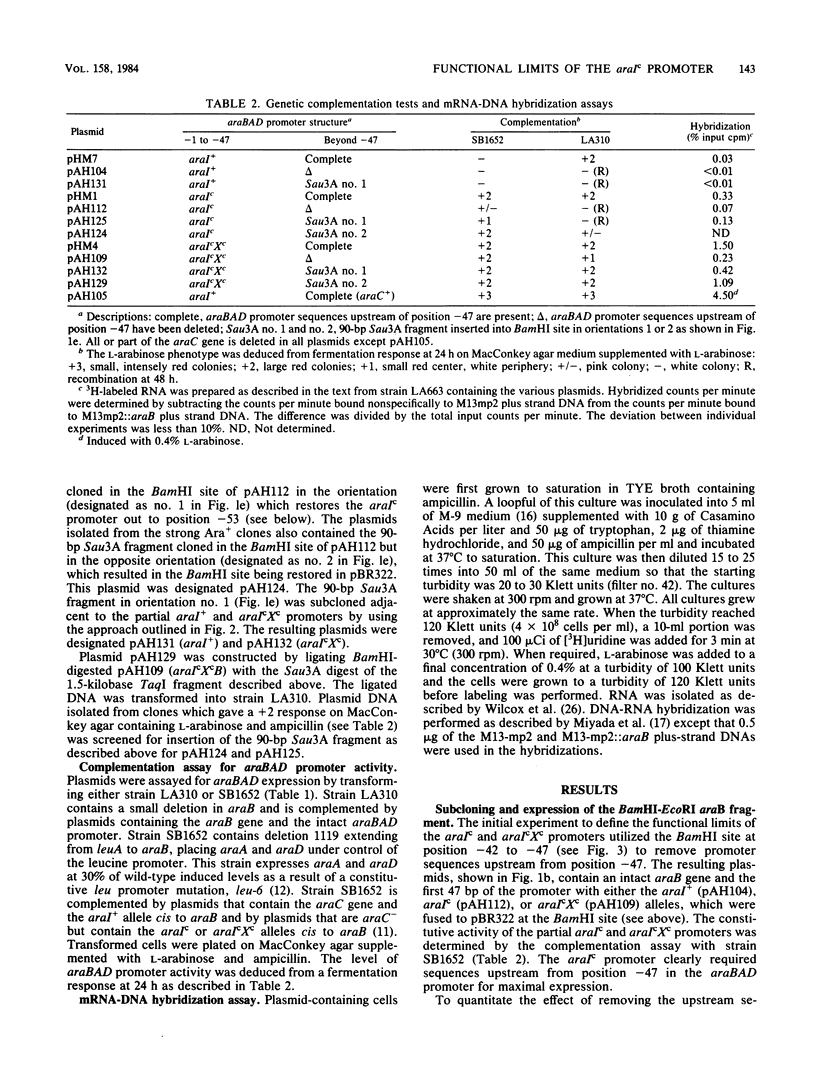
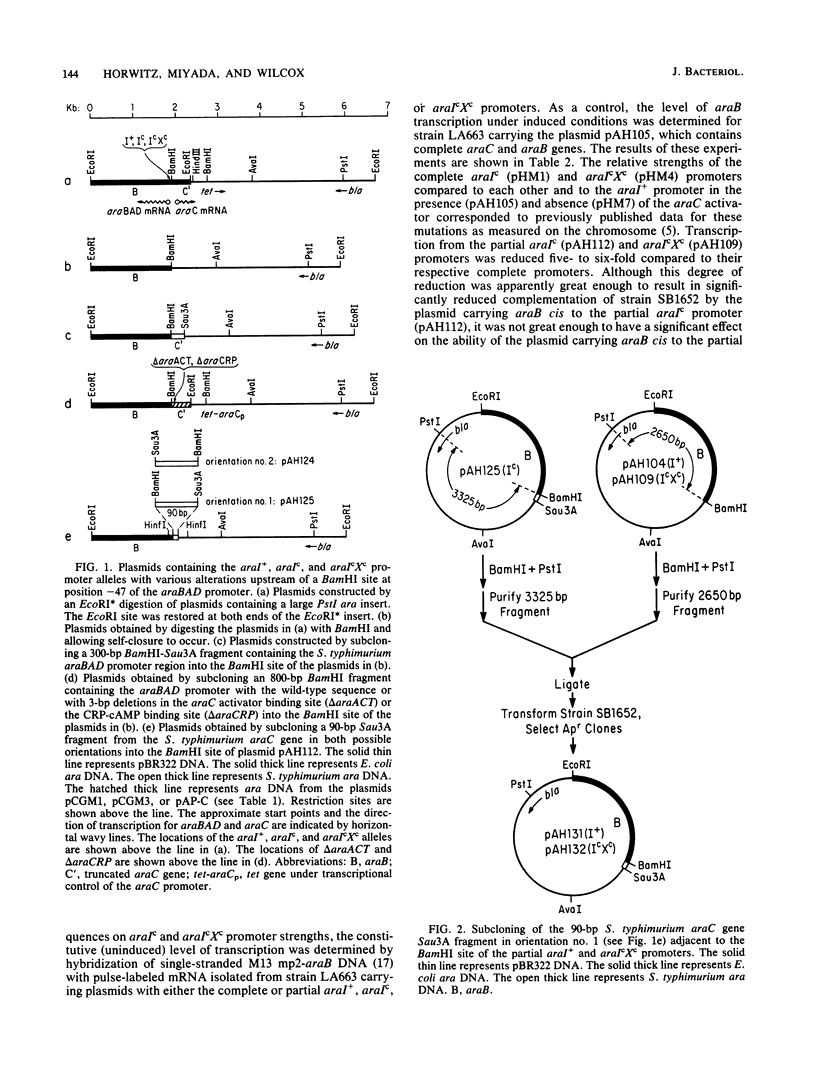
The araBAD promoter is defined, in part, by two types of cis-acting constitutive mutations, araIc at position -35 and araXc at position -10. Subcloning experiments demonstrated that the araIc and araIcXc promoters require DNA sequence information out to position -53 to -56 for maximum constitutive expression. This is 8 to 10 base pairs more DNA than is generally thought to be necessary for RNA polymerase interaction. The -53 to -56 region is required for glucose repression, suggesting that an additional factor interacts in this region and is necessary for maximum expression.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bass R., Heffernan L., Sweadner K., Englesberg E. The site for catabolite deactivation in the L-arabinose BAD operon in Escherichia coli B/r. Arch Microbiol. 1976 Oct 11;110(1):135–143. doi: 10.1007/BF00416978. [DOI] [PubMed] [Google Scholar]
- Cass L. G., Horwitz A. H., Miyada C. G., Greenfield L., Wilcox G. The araC regulatory gene mRNA contains a leader sequence. Mol Gen Genet. 1980;180(1):219–226. doi: 10.1007/BF00267373. [DOI] [PubMed] [Google Scholar]
- Cass L. G., Horwitz A. H., Wilcox G. The araIc mutation in Escherichia coli B/r. J Bacteriol. 1981 Jun;146(3):1098–1105. doi: 10.1128/jb.146.3.1098-1105.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colomé J., Wilcox G., Englesberg E. Constitutive mutations in the controlling site region of the araBAD operon of Escherichia coli B/r that decrease sensitivity to catabolite repression. J Bacteriol. 1977 Feb;129(2):948–958. doi: 10.1128/jb.129.2.948-958.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englesberg E., Sheppard D., Squires C., Meronk F., Jr An analysis of "revertants" of a deletion mutant in the C gene of the L-arabinose gene complex in Escherichia coli B-r: isolation of initiator constitutive mutants (Ic). J Mol Biol. 1969 Jul 28;43(2):281–298. doi: 10.1016/0022-2836(69)90268-x. [DOI] [PubMed] [Google Scholar]
- Englesberg E., Wilcox G. Regulation: positive control. Annu Rev Genet. 1974;8:219–242. doi: 10.1146/annurev.ge.08.120174.001251. [DOI] [PubMed] [Google Scholar]
- Gielow L., Largen M., Englesberg E. Initiator constitutive mutants of the L-arabinose operon (OIBAD) of Escherichia coli B/r. Genetics. 1971 Nov;69(3):289–302. doi: 10.1093/genetics/69.3.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz A. H., Heffernan L., Morandi C., Lee J. H., Timko J., Wilcox G. DNA sequence of the araBAD-araC controlling region in Salmonella typhimurium LT2. Gene. 1981 Sep;14(4):309–319. doi: 10.1016/0378-1119(81)90163-3. [DOI] [PubMed] [Google Scholar]
- Horwitz A. H., Morandi C., Wilcox G. Deoxyribonucleic acid sequence of araBAD promoter mutants of Escherichia coli. J Bacteriol. 1980 May;142(2):659–667. doi: 10.1128/jb.142.2.659-667.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler D. P., Englesberg E. Arabinose-leucine deletion mutants of Escherichia coli B-r. J Bacteriol. 1969 Jun;98(3):1159–1169. doi: 10.1128/jb.98.3.1159-1169.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Grice S. F., Matzura H. Binding of RNA polymerase and the catabolite gene activator protein within the cat promoter in Escherichia coli. J Mol Biol. 1981 Aug 5;150(2):185–196. doi: 10.1016/0022-2836(81)90448-4. [DOI] [PubMed] [Google Scholar]
- Lee N. L., Gielow W. O., Wallace R. G. Mechanism of araC autoregulation and the domains of two overlapping promoters, Pc and PBAD, in the L-arabinose regulatory region of Escherichia coli. Proc Natl Acad Sci U S A. 1981 Feb;78(2):752–756. doi: 10.1073/pnas.78.2.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyada C. G., Soberón X., Itakura K., Wilcox G. The use of synthetic oligodeoxyribonucleotides to produce specific deletions in the araBAD promoter of Escherichia coli B/r. Gene. 1982 Feb;17(2):167–177. doi: 10.1016/0378-1119(82)90070-1. [DOI] [PubMed] [Google Scholar]
- Norgard M. V., Emigholz K., Monahan J. J. Increased amplification of pBR322 plasmid deoxyribonucleic acid in Escherichia coli K-12 strains RR1 and chi1776 grown in the presence of high concentrations of nucleoside. J Bacteriol. 1979 Apr;138(1):270–272. doi: 10.1128/jb.138.1.270-272.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden S., Haggerty D., Stoner C. M., Kolodrubetz D., Schleif R. The Escherichia coli L-arabinose operon: binding sites of the regulatory proteins and a mechanism of positive and negative regulation. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3346–3350. doi: 10.1073/pnas.77.6.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queen C., Rosenberg M. A promoter of pBR322 activated by cAMP receptor protein. Nucleic Acids Res. 1981 Jul 24;9(14):3365–3377. doi: 10.1093/nar/9.14.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T., O'Neill M., de Crombrugghe B. Interaction site of Escherichia coli cyclic AMP receptor protein on DNA of galactose operon promoters. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5090–5094. doi: 10.1073/pnas.76.10.5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox G., Singer J., Heffernan L. Deoxyribonucleic acid-ribonucleic acid hybridization studies on the L-Arabinose operon of Escherichia coli B-r. J Bacteriol. 1971 Oct;108(1):1–4. doi: 10.1128/jb.108.1.1-4.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]


