The free energy of partitioning an amino acid side chain from water into the cell membrane is one of the critical parameters for understanding and predicting membrane protein stability, and understanding membrane protein function. Transmembrane segments are generally very hydrophobic, but may contain hydrophilic residues that are important for the structure or function of the protein. Experimental and theoretical studies have shown that the presence of polar residues, such as Asn, can lead to the formation of helical aggregates (Stockner et al., 2004; Tatko et al., 2006). The crystal structures of the voltage-gated potassium channels KvAP (Jiang et al., 2003a) and Kv1.2 (Long et al., 2005) have caused vigorous debate in the ion channel community as some models proposed based on the crystal structures would have the arginine gating charges exposed to the lipid environment (Jiang et al., 2003b). After the publication of the KvAP crystal structure, it was argued that it was next to impossible to put an arginine in a lipid-exposed environment, and that the activation energy for such a model would be far too high to be realistic (Grabe et al., 2004). However, a recent experimental study has shown that the S4 segment of KvAP, which contains the gating charges, is able to insert into the membrane as a marginally stable transmembrane helix (Hessa et al., 2005b).
We currently have a limited understanding of the partitioning behavior of amino acids into lipid bilayers. Numerous experimental scales have been derived using a variety of model systems and the results of such experiments have proven very useful in the prediction of membrane protein stability. The Perspectives from White, Wolfenden, and von Heijne in this issue outline several experimental scales and discuss their importance for understanding the membrane environment. Molecular dynamics (MD) computer simulations provide a complementary view of side chain partitioning, providing a level of detail that is not accessible to experiment. We have recently performed a systematic set of calculations (unpublished data) on the distributions of 17 of 20 amino acids (Pro, Gly, and His excluded). Here, we will compare the results of these simulations to several experimental scales.
Computational Results
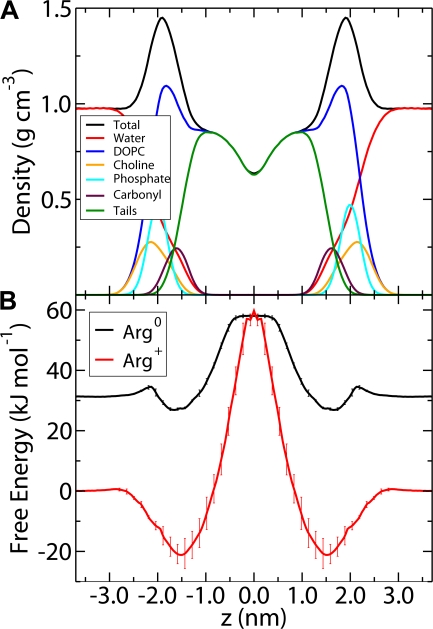
We will focus on the results of our recent work. Following Wolfenden's experimental studies (see Perspective in this issue), we simulated small molecule analogues of the amino acid side chains. The side chains were truncated at the β-carbon with the α-carbon replaced by a proton. For example, phenylalanine becomes toluene, and isoleucine becomes butane. For simplicity, we will refer to the compounds by the three-letter code of the corresponding amino acid. Simulations were performed on a system containing 64 lipid molecules, 2,804 water molecules, and two side chains. An umbrella sampling protocol was employed to determine the potential of mean force for the side chain in the lipid bilayer. A total of 37 simulations were performed for each residue, with each simulation having a minimum length of 30 ns, for a total of 1.1 μs per residue. For some residues, such as Trp and Arg, the simulations were extended up to 80 ns in order to improve the accuracy of the calculation. Based on the calculated free energy profiles we have determined two scales: one for the center of the membrane and one for the interfacial region, summarized in Table I. Fig. 1 A shows a partial density profile, indicating the location of various lipid functional groups; Fig. 1 B shows the potential of mean force (PMF) for the charged and neutral forms of Arg.
TABLE I.
Calculated Side Chain Transfer Free Energies
| Residue | Water to DOPC Interface (kJ/mol) |
Water to Center of DOPC Membrane (kJ/mol) |
|---|---|---|
| Leu | −14.1 | −15.2 |
| Ile | −20.6 | −22.1 |
| Val | −12.2 | −13.8 |
| Phe | −14.9 | −12.8 |
| Ala | −6.8 | −8.4 |
| Trp | −21.6 | −4.9 |
| Met | −10.5 | −4.4 |
| Cys | −6.6 | −3.4 |
| Tyr | −14.0 | 6.6 |
| Thr | −4.2 | 13.9 |
| Ser | −0.7 | 15.8 |
| Gln | −8.9 | 20.2 |
| Lys | −18.6a | 19.9b |
| Asn | −6.5 | 23.9 |
| Glu | −1.68b | 21.1b |
| Asp | 1.6b | 31.0b |
| Arg | −21.2a | 58.1c |
Residue is charged at this location.
Residue is neutral at this location.
Charged and neutral forms are equally likely.
Figure 1.
Partial density profiles and sample PMFs. (A) Partial density profiles of various lipid functional groups to serve as a reference for the location of different depths in the bilayer. (B) PMFs of the charged and neutral species of Arg. The neutral form has been shifted by the free energy to deprotonate an Arg in bulk water.
The aliphatic residues (Ala, Val, Leu, Ile) partition favorably to both the interfacial region and the center of the bilayer. Partitioning to the interface region is almost as favorable as the center of the bilayer; it is reduced slightly due to the increase in density and lipid chain order at the interface (MacCallum and Tieleman, 2006). Alanine is the least hydrophobic of the aliphatic residues, while Ile is the most hydrophobic.
Cys and Met partition favorably to both the interface and the center of the bilayer. The partitioning of Met is more favorable than Cys, at both the interface and the center of the bilayer. Unlike the aliphatic residues, however, the interface is favored over the center of the bilayer for both Cys and Met. In terms of partitioning, the small molecule analogues of both Cys and Met behave similarly to Trp (see below).
Phe partitions similarly to the aliphatic residues, with nearly equal free energies in the interface and the center of the bilayer. Based on partitioning into the center of the bilayer, Phe is the fourth most hydrophobic residue, behind Ile, Leu, and Val. Both Trp and Tyr display deep interfacial wells in the vicinity of the carbonyl groups. Partitioning of Tyr to the center of the bilayer has an unfavorable free energy of +6 kJ/mol due to the loss of hydration for the polar hydroxyl group. In terms of partitioning, Tyr behaves most similarly to the polar residues (see below). Trp, being less polar than Tyr, still has a favorable free energy for transfer to the center of the bilayer at −4.9 kJ/mol. Trp is often regarded as being a very hydrophobic residue; however these results, and those of Wolfenden, show that Ala has a comparable free energy at the center of the membrane. The preference of Trp and Tyr for the interface in known membrane protein structures (Yau et al., 1998) correlates with the strong interfacial partitioning shown here.
A recent paper presented a very detailed study on the partitioning of analogues of Phe (benzene) and Trp (indole) into a POPC bilayer (Norman and Nymeyer, 2006). Phe was found to partition equally to both the center of the membrane and the interface, while Trp partitioned preferentially to the interface, in agreement with our findings. Norman and Nymeyer showed that the interfacial preference of Trp was due to a combination of weak electrostatic interactions and an otherwise hydrophobic molecule. Presumably, similar electrostatic interactions may be responsible for the interfacial partitioning of Tyr.
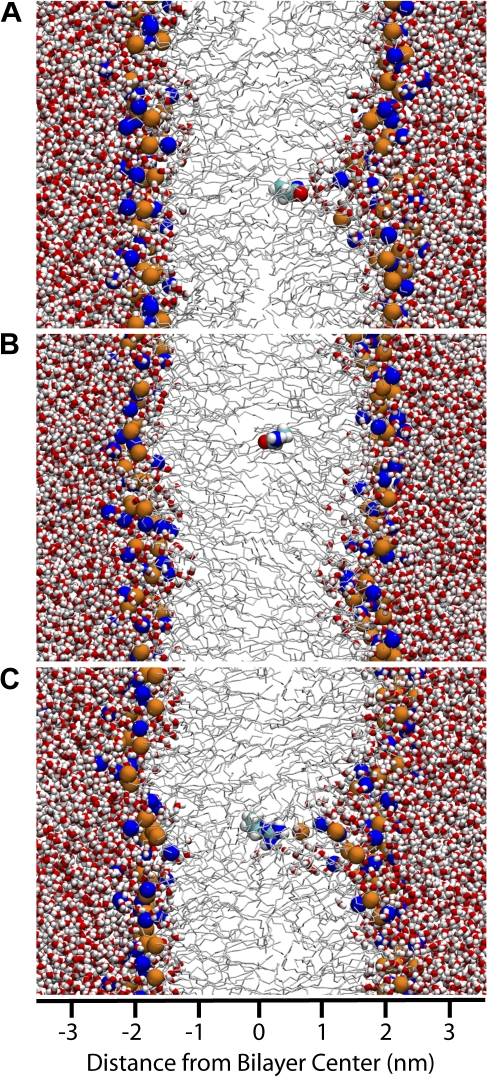
The PMF of the neutral form of Arg (Arg0 in Fig. 1 B) is representative of the polar residues (Asn, Gln, Ser, and Thr), which display an interfacial free energy minimum in the vicinity of the carbonyl groups, with depths ranging from −8.9 kJ/mol for Gln to −0.7 kJ/mol for Ser. The free energy increases rapidly as the side chain is moved closer to the center of bilayer until, at around 0.5 nm from the center, the free energy plateaus (unpublished data). Partitioning to the center of the membrane is unfavorable for all of the polar residues, ranging from 23.9 kJ/mol for Asn to 13.9 kJ/mol for Thr. The plateau in free energy at the center of the bilayer correlates well with the occurrence of water defects in the membrane structure. At positions close to the membrane interface, the polar residue is hydrated by a water defect (Fig. 2 A). As the residue moves toward the center of the membrane, the size of the defect increases and becomes more energetically unfavorable. At some point, the cost of creating the defect outweighs the energy gained by hydrating the polar side chain and the water defect dissipates, leaving an isolated side chain in the central region of the bilayer (Fig. 2 B).
Figure 2.
Selected snapshots of the simulated systems. Red and white sticks are water; DOPC phosphate groups are orange spheres; DOPC choline groups are blue spheres; the remainder of DOPC molecule is rendered as light gray lines. The space filling molecule in the center of the bilayer is the side chain of interest, colored as cyan (carbon), white (hydrogen), blue (nitrogen), and red (oxygen). (A) Snapshot of Gln at 0.4 nm from the center of the bilayer. The residue is stabilized by a large water defect. (B) Snapshot of Gln at 0.3 nm from the center of the bilayer. The water defect is no longer stable. (C) Snapshot of Arg at the center of the membrane. The water defect is stable over the length of the simulation (80 ns). Note that the water defect is partially lined by a lipid headgroup.
Johansson and Lindahl (2006) have performed an extensive set of simulations on substituted hydrophobic transmembrane peptides. When polar or charged residues were substituted near the center of the peptide, similar water defects were formed that hydrated the polar side chain. Although they did not calculate the free energies of partitioning, they observed a correlation between side chain hydration and the biological (Hessa et al., 2005a) and water-octanol (Wimley et al., 1996) hydrophobicity scales from White and coworkers.
The PMF of positively charged Arg (Fig. 1 B) is representative of the charged forms of the ionizable residues. The free energy increases rapidly in the acyl chain regions and peaks at the center of the membrane. Large water defects are observed, penetrating all the way to the center of the membrane (Fig. 2 C). However, the charge state of the ionizable residues in a lipid bilayer is not obvious, as the cost of partitioning a charge into a hydrophobic environment is balanced against the cost of neutralizing the residue. To investigate the partitioning of the charged residues, we have performed simulations of both the ionized and neutral forms of Arg, Lys, Glu, and Asp. The PMF for the neutral form can then be shifted by the free energy required to neutralize the molecule at pH 7.0 relative to the charged form. This allows us to determine where in the membrane each residue is ionized, and to obtain a composite free energy profile that reflects the ionization state. Fig. 1 B shows the PMF for both the charged and neutral forms of Arg. The PMFs for the other charged residues are similar. Arg is the least favorable residue at the center of the membrane, followed by Asp, Glu, and Lys. The acidic residues, Asp and Glu, are ionized in bulk water and near the choline groups, but are neutral in the region from the carbonyl group to the center of the bilayer. The neutral forms are stabilized (relative to bulk hydrocarbon) by a water defect until they are ∼0.3 nm from the bilayer center where the water defect dissipates. The basic residues remain ionized until deeper into the bilayer. For Lys, the crossover point occurs at ∼0.4 nm from the center of the bilayer, well within the acyl chain region. In contrast to the other charged residues, for Arg the ionized form is more stable than the neutral from until the very center of the membrane where the neutral and ionized forms are nearly equal in free energy at 58 and 59 kJ/mol, respectively. This raises the very interesting possibility that arginine could remain ionized even at the center of the membrane (Fig. 2 C), at a much lower free energy cost than expected based on continuum arguments because of the presence of hydrating water.
A recent paper presents a very detailed set of calculations on the partitioning of a polyleucine helix containing a central positively charged arginine residue into a DPPC bilayer (Dorairaj and Allen, 2007). They report a free energy barrier of 71 kJ/mol for arginine at the center of the membrane. We calculate a value of 58 kJ/mol, which is significantly lower. However, because the Arg in the Dorairaj study is in a poly-Leu helix, the relevant quantity to compare to is the free energy of transferring an Arg from water to the center of the membrane plus the free energy of transferring a Leu from the center of the membrane to water. Using this scheme, we calculate a value of 58.0 + 15.2 = 73 kJ/mol; in good agreement with the results obtained by Dorairaj and Allen using a different force field. They also observe very large water defects similar to those observed in our study (Fig. 1 C). Other recent simulations have also observed similar water defects in transmembrane helices containing Arg residues (Freites et al., 2005; Johansson and Lindahl, 2006). Taken together, these results point to the importance of the flexible and deformable nature of the bilayer. Continuum electrostatics calculations of a transmembrane helix containing arginine at the center of the membrane give a free energy of up to 170 kJ/mol, depending on the assumptions made (Dorairaj and Allen, 2007); a value which is double that obtained from all-atom simulations. The lipid bilayer should not be understood as a static, low dielectric slab, but rather as a complex and rather easily deformable system.
Dorairaj and Allen (2007) simulated only the ionized form of arginine. The free energy difference between the neutral and ionized forms is known in bulk water, but is not known in the membrane environment because the pKa shifts by an unknown amount. To overcome this issue, we have performed simulations of both the charged and neutral species. Based on our calculations, lipid-exposed Lys, Asp, and Glu residues would be present as neutral species if lipid exposed at the center of a DOPC membrane. On the other hand, our results suggest that the ionized and neutral forms of Arg are nearly equal in free energy, and thus equal in population at the center of the membrane, at least the DOPC model membrane in our simulations.
Comparison to Experiment
Cyclohexane.
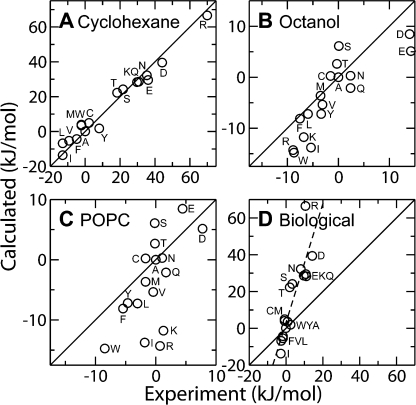
Fig. 3 A shows the correlation between the experimental water to cyclohexane transfer free energy (Radzicka and Wolfenden, 1988), and our calculated values for transferring the side chain from water to the center of the membrane. Although the experiments do not involve lipid bilayers, we compare to them for two reasons. First, the experiments measure the partitioning of the same small molecule analogues of the amino acid side chains that we use in our calculations, allowing a more direct comparison than with the peptide-based systems discussed below. Second, cyclohexane is a very simple hydrophobic phase, and it is clear what the local environment of the side chain is, which is not the case in the other experimental systems. The solid line in Fig. 3 A denotes perfect agreement between experiment and calculation, and is not a best-fit line. There is a very strong correlation (correlation coefficient 0.99) between the experimental and calculated results across a wide range of free energies (−20 to +60 kJ/mol). Good agreement between the center of the membrane and a bulk alkane, such as cyclohexane, should be expected as both phases present a pure alkane environment of similar density. In particular, the free energies for the charged residues are in very good agreement, which supports the conclusions on protonation state presented earlier.
Figure 3.
Comparison of calculated results (vertical axis) to experimental results (horizontal axis). Residues are indicated by a circle and labeled with the corresponding single letter amino acid code. Solid lines indicate perfect agreement between calculation and experiment. In all cases, the experimental and calculated results have been normalized to the free energy of Ala. (A) Comparison of the calculated free energy at the center of the membrane and the experimental water-cyclohexane transfer free energy (Radzicka and Wolfenden, 1988). (B) Correlation between the calculated free energy at the interface and the experimental octanol-water transfer free energy. Experimental values are taken from the ΔGWLXLL column of Table I in Wimley et al. (1996), normalized to the value of Ala. (C) Comparison of the calculated free energy at the interface and the experimental DOPC-water transfer free energy. Experimental data is taken from from the ΔGWLXLL column of Table I in Wimley and White (1996). (D) Comparison between the calculated free energy at the center of the membrane and biological free energy scale based on measurements using the Lep system (Hessa et al., 2005a). The dashed line is a linear regression to the data and has a slope of 3.4.
Octanol Pentapeptide Scale
The correlation between the partitioning of a series of AcWLxLL peptide between water and octanol (Wimley et al., 1996), and our calculated water to interface scale is shown in Fig. 3 B. The correlation between the scales is high with a correlation coefficient of 0.84. Even the partitioning of the charged side chains is in relatively good agreement between the scales. The agreement between the octanol scale and our calculated values at the center of the membrane is poor (not depicted), supporting the suggestion that octanol is a good mimetic for the membrane interface. Water saturated octanol is inhomogeneous on the microscopic scale and presents a mix of polar environments composed of hydroxyl groups and water, hydrophobic environments containing the alkane tails, as well as the interface between them (MacCallum and Tieleman, 2002). Wimley et al. attempted to correct their measured free energies to account for occlusion effects and changes in solvent- accessible surface area in their measurements. However, we found that our calculated scale agreed much better with their raw experimental data than with the corrected values (not depicted).
POPC Pentapeptide Scale
Fig. 3 C shows the correlation between the partitioning of AcWLxLL peptides into POPC vesicles (Wimley and White, 1996), and our calculated interfacial scale. The correlation between the two scales is relatively low at 0.61; however Lys, Arg, and Ile are clear outliers. Excluding these residues improves the correlation to 0.85. We believe that the three outliers can be explained by the fact that we have used small molecule analogues of the side chains, rather than the pentapeptides used in the experiment. The most favorable orientation for the small molecule analogues of Arg and Lys is to have the charged group buried in the carbonyl region of the lipid bilayer, while the alkane tail partitions into the acyl chains of the lipid. In contrast, that orientation is not easily available for side chains within the peptide due to constraints imposed by the rest of the peptide (Aliste and Tieleman, 2005). Thus, it should be expected that the free energy in the peptide system would be higher than calculated from small molecule analogues. While this effect applies to all residues, it should be strongest for Arg and Lys as they are the most amphiphilic of all of the compounds studied here. The poor correlation of Ile may also be explained by differences between the small molecule analogues we have simulated and the peptides used in the experiment. The analogue of Ile we have used (butane) is linear, that of Leu (isobutane) is branched, while in the peptide both Ile and Leu are branched. Partitioning of a linear alkane should be more favorable than for a branched one. One therefore would expect that the free energy for Leu should be relatively close to experiment, while that of Ile will be too low, which is indeed the case.
Biological Scale
A recent collaboration between the White and von Heijne laboratories attempted to define the “biological” hydrophobicity scale (Hessa et al., 2005a). They challenged the Sec61 translocon with a variety of constructs based on modification of the leader peptidase sequence. By determining the fraction of the modified helix that was inserted into the membrane, they were able to determine the pseudo free energy for placing each side chain into the center of the membrane. The correlation between the biological scale and our calculated scale for the center of the membrane is shown in Fig. 3 D. The correlation coefficient is 0.87. However, the slope of the best-fit line is nearly 3.4, meaning that the calculated values vary much more than the experimental values. For example, Hessa et al. report a value of 10.5 kJ/mol for arginine (normalized to the value for Ala), while we obtain a value of 66 kJ/mol. Similarly, our results indicate that the hydrophobic residues are much more favorable in the center of the membrane compared with experiment. Interestingly, the results from the Wolfenden laboratory (Radzika and Wolfenden, 1998) display the same trend; the correlation coefficient is high, but the slope is much greater than unity. There are a number of possible reasons for this discrepancy. First, the experimental system is very complicated and it is not clear what the local environment of the side chain is. It has been suggested that the more polar and charged residues may be buried in contacts with the Sec61 translocon (Hessa et al., 2005a). Alternatively, as the Lep protein contains two other transmembrane helices, its residues may be buried within protein–protein contacts and may not be lipid exposed. Second, we have studied small molecule analogues, while the experimental system uses peptides. The residue of interest may be partially occluded by neighboring residues or the peptide backbone may stabilize the formation of water defects leading to lower free energies for the charged and polar side chains. Third, the transmembrane segment used in the experiments may not be strongly anchored, leading to the possibility that the entire helix is translated so that the polar or charged residue has a favorable environment, as suggested by the calculations of Dorairaj and Allen (2007). It may be cheaper to translate the entire helix so that the arginine residue was closer to the water phase, than to have the arginine in the center of the membrane with a large water defect. One can see that the primary limitation of the biological scale is that the local chemical environment of the substituted side chain is unknown, as also eluded to by von Heijne in his Perspective in this issue. Based on the good agreement between our calculated results and the results of Radzika and Wolfenden, and the large discrepancy between those results and the biological scale, we can conclude that the biological scale probably does not reflect the free energy for a lipid-exposed side chain. However, at this point we do not know exactly what environment the biological scale represents. Interestingly, the correlation coefficients between both our calculated scale and the experimental water-cyclohexane scale with the biological scale are higher than for either the octanol or interfacial scales of the White laboratory.
Implications for Voltage Gating in Ion Channels
At this point in time, computer simulations have not resolved the mechanism of ion channel gating. However, the formation of water defects when an Arg is placed in the membrane has emerged as a consistent feature of a number of recent simulations. Freites et al. have shown that if S4 is inserted in a transmembrane orientation, large water defects are formed, although they did not demonstrate the thermodynamic stability of this configuration. More recent simulations using a coarse grained model have shown that the transmembrane and surface bound orientations of S4 appear to be similar in free energy (Bond and Sansom, 2007). Our results, along with those of Dorairaj and Allen, show that the cost of transferring an Arg from water to the center of the membrane is relatively high. However, this high cost can be offset by the favorable free energy for burying nonpolar residues such as Leu in the hydrocarbon core. Ignoring backbone contributions, the transfer of four Leu residues from water to the center of the membrane should be enough to overcome the unfavorable free energy of Arg at the center of the bilayer. It is also important to note that the relevant barrier for gating is not the free energy change from bulk water to the center of the membrane. Instead, if the Arg residues already reside partially in the membrane environment, as in the Kv1.2 crystal structure (Long et al., 2005), the barrier is the free energy from that location to the center of the membrane. Finally, the formation of a water defect suggests that the free energy may be nonadditive with respect to the number of Arg residues. That is, while the free energy for a single Arg at the center of the membrane is 60 kJ/mol, it is likely that the free energy for four Arg residues is <240 kJ/mol, possibly significantly less. If several Arg residues are in close proximity, as in S4, the free energy is likely determined by the deepest penetrating residue, as the remaining residues are able to “piggyback” on the water defect formed by the deepest residue at little cost.
Concluding Remarks
Computer simulations have enhanced our understanding of the partitioning of amino acids into lipid bilayers as they provide a level of detail that is not accessible to experiment. In particular, the observation of water defects that stabilize polar and charged residues in the hydrocarbon core was unexpected. The appearance of these water defects is robust across simulations from a number of different groups, each using different models and force fields. The presence of water defects dramatically alters the free energy profiles, and reinforces the point that membranes should not be viewed as static low dielectric slabs.
In general, the agreement between the simulated distributions and the measured free energies is good. The calculated interfacial scale correlates well with both the octanol and POPC interfacial scales. The calculated values at the center of the bilayer agree very well with measured water-cyclohexane transfer free energies. There is also a strong correlation between the calculated values at the center of the bilayer and the biological hydrophobicity scale, although in this case the calculated values vary by ∼3.4 times more than the experimental value.
The primary limitation of the experimental studies is that it is very difficult to control or measure the local environment of the side chain. Thus, the measured free energies may be more difficult to interpret than it first appears. Likewise, simulations are limited by computational cost, force field accuracy, and complexity. As computer power increases and molecular force fields become more accurate, the complexity of systems that can be studied will also improve and results will become more quantitative.
Acknowledgments
This work is supported by the Natural Sciences and Engineering Research Council (NSERC). J.L. MacCallum is supported by studentships from NSERC, Alberta Ingenuity, and the Killam Trust. D.P. Tieleman is an AHFMR Senior Scholar and CIHR New Investigator.
Abbreviation used in this paper: PMF, potential of mean force.
References
- Aliste, M.P., and D.P. Tieleman. 2005. Computer simulation of partitioning of ten pentapeptides Ace-WLXLL at the cyclohexane/water and phospholipid/water interfaces. BMC Biochem. 6:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond, P.J., and M.S.P. Sansom. 2007. Bilayer deformation by the Kv voltage sensor domain revealed by self-assembly simulations. Proc. Natl. Acad. Sci. USA. 104:2631–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorairaj, S., and T.W. Allen. 2007. On the thermodynamic stability of a charged arginine sidechain in a transmembrane helix. Proc. Natl. Acad. Sci. USA. In press. [DOI] [PMC free article] [PubMed]
- Freites, J.A., D.J. Tobias, G. von Heijne, and S.H. White. 2005. Interface connections of a transmembrane voltage sensor. Proc. Natl. Acad. Sci. USA. 102:15059–15064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabe, M., H. Lecar, Y.N. Jan, and L.Y. Jan. 2004. A quantitative assessment of models for voltage-dependent gating of ion channels. Proc. Natl. Acad. Sci. USA. 101:17640–17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessa, T., H. Kim, K. Bihlmaler, C. Lundin, J. Boekel, H. Andersson, I. Nilsson, S.H. White, and G. von Heijne. 2005. a. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 433:377–381. [DOI] [PubMed] [Google Scholar]
- Hessa, T., S.H. White, and G. von Heijne. 2005. b. Membrane insertion of a potassium-channel voltage sensor. Science. 307:1427. [DOI] [PubMed] [Google Scholar]
- Jiang, Y., A. Lee, J.Y. Chen, V. Ruta, B.T. Chait, and R. MacKinnon. 2003. a. X-ray structure of a voltage-dependent K+ channel. Nature. 423:33–41. [DOI] [PubMed] [Google Scholar]
- Jiang, Y., V. Ruta, J.Y. Chen, A. Lee, and R. MacKinnon. 2003. b. The principle of gating charge movement in a voltage-dependent K+ channel. Nature. 423:42–48. [DOI] [PubMed] [Google Scholar]
- Johansson, A.C.V., and E. Lindahl. 2006. Amino-acid solvation structure in transmembrane helices from molecular dynamics simulations. Biophys. J. 91:4450–4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, S.B., E.B. Campbell, and R. MacKinnon. 2005. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 309:897–903. [DOI] [PubMed] [Google Scholar]
- MacCallum, J.L., and D.P. Tieleman. 2002. Structure of neat and hydrated 1-octanol from computer simulations. J. Am. Chem. Soc. 124:15085–15093. [DOI] [PubMed] [Google Scholar]
- MacCallum, J.L., and D.P. Tieleman. 2006. Computer simulation of the distribution of hexane in a lipid bilayer: spatially resolved free energy, entropy, and enthalpy profiles. J. Am. Chem. Soc. 128:125–130. [DOI] [PubMed] [Google Scholar]
- Norman, K.E., and H. Nymeyer. 2006. Indole localization in lipid membranes revealed by molecular simulation. Biophys. J. 91:2046–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radzicka, A., and R. Wolfenden. 1988. Comparing the polarities of the amino acids: side-chain distribution coefficients between the vapor phase, cyclohexane, 1-octanol, and neutral aqueous solution. Biochemistry. 27:1664–1670. [Google Scholar]
- Stockner, T., W.L. Ash, J.L. MacCallum, and D.P. Tieleman. 2004. Direct simulation of transmembrane helix association: role of asparagines. Biophys. J. 87:1650–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatko, C.D., V. Nanda, J.D. Lear, and W.F. DeGrado. 2006. Polar networks control oligomeric assembly in membranes. J. Am. Chem. Soc. 128:4170–4171. [DOI] [PubMed] [Google Scholar]
- Wimley, W.C., T.P. Creamer, and S.H. White. 1996. Solvation energies of amino acid side chains and backbone in a family of host-guest pentapeptides. Biochemistry. 35:5109–5124. [DOI] [PubMed] [Google Scholar]
- Wimley, W.C., and S.H. White. 1996. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 3:842–848. [DOI] [PubMed] [Google Scholar]
- Yau, W.-M., W.C. Wimley, K. Gawrisch, and S.H. White. 1998. The preference of tryptophan for membrane interfaces. Biochemistry. 37:14713–14718. [DOI] [PubMed] [Google Scholar]