Introduction
While many generalizations about membrane protein assembly and structure have been formulated over the years, most recently based on the rapidly growing database of high-resolution three-dimensional membrane protein structures (White, 2004), a quantitative understanding of the underlying principles is mostly lacking, at least for in vivo conditions. To address this issue, we have developed an experimental system based on in vitro translation of model membrane proteins in the presence of dog pancreas rough microsomes that makes it possible to measure the efficiency with which different natural or designed polypeptide segments insert into the ER membrane under conditions approximating the in vivo situation (Sääf et al., 1998; Hessa et al., 2005a,b; Meindl-Beinker et al., 2006). Here, I will first summarize the basic mechanism of membrane protein insertion in the ER and will then review the main results that we have obtained so far using the microsomal in vitro system. From a conceptual point of view, the data we have generated point to protein–lipid interactions as providing the main driving force for membrane integration, despite the fact that integration is mediated by a very complex molecular machine: the Sec61 translocon. Finally, I will discuss what possible bearing our results might have on an issue close to the hearts of many molecular physiologists: the workings of ion channel voltage-sensor domains.
Membrane Protein Insertion into the ER
While the insertion of a hydrophobic membrane protein into a biological membrane might appear to be a simple question of partitioning from water into a lipid bilayer, this is not how it works in a cell. Above all, the strongly apolar stretches of polypeptide that will eventually form the transmembrane α helices must be prevented from aggregating before they reach their proper destination; therefore, nearly all membrane proteins are synthesized on membrane-bound ribosomes and delivered directly into the membrane with the aid of a so-called translocon, an integral membrane–protein complex that can guide polypeptides both across and into the surrounding lipid bilayer.
The best characterized translocon is found in the ER membrane of mammalian cells, the so-called Sec61 complex (Alder and Johnson, 2004). Ribosomes translating mRNAs encoding secretory and membrane proteins are targeted to the Sec61 translocon with the help of the signal recognition particle (SRP), a protein–RNA particle that recognizes hydrophobic signal sequences as they emerge from the ribosome. In a process orchestrated by mutually dependent GTPase activities on the SRP and the translocon-bound SRP receptor, the ribosome is positioned at the top of the translocon channel, and the nascent polypeptide chain is fed directly from the polypeptide-conducting tunnel in the large ribosomal subunit into the translocon.
From the point of view of membrane protein biogenesis, the most important aspect of this process is how the ribosome–translocon complex manages to recognize the incipient transmembrane helices in the nascent polypeptide and allows them to insert into the lipid bilayer rather than being fully translocated across the membrane into the lumen of the ER. From the only high-resolution structure available, a monomeric Sec61αβγ complex from the archean Methanococcus jannaschii (van den Berg et al., 2004), and from lower-resolution single-particle electron microscopy structures (Beckmann et al., 2001; Mitra et al., 2005), it appears that transmembrane segments leave the channel through a lateral gate in the channel wall that opens sideways toward the bilayer. The precise molecular environment and the mechanistic details behind the recognition of transmembrane helices are still obscure, however.
The “Biological” Hydrophobicity Scale
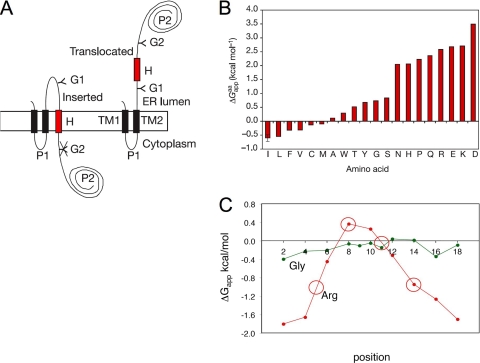
Our approach to the question of what features of the nascent chain determine the membrane insertion propensity of polypeptide segments in vivo has been “substrate engineering,” i.e., we have set up an assay in which the Sec61 translocon is challenged by large sets of designed, more or less hydrophobic, test segments (H segments) engineered into a model membrane protein, Fig. 1 A. To be able to measure the degree of membrane insertion of the H segments, two acceptor sites for N-linked glycosylation have been inserted into the model protein, one on each side of the H-segment. Since the oligosaccharide transferase enzyme that catalyzes the addition of glycans to the nascent polypeptide is located in the lumen of the ER, the degree of membrane insertion of a given H-segment can be quite accurately measured as the quotient between the amount of singly glycosylated (f 1) and doubly (f 2) glycosylated protein, and expressed as an apparent free energy difference (ΔGapp = −RT ln(f 1/f 2)) between the inserted and noninserted states (“apparent,” because we do not know if the process has time to reach equilibrium before the potential transmembrane segment has been pushed out of the translocon by the growing nascent chain). The assay can be performed both in vivo, and, more easily, in vitro by using a coupled transcription–translation system supplanted by dog pancreas rough microsomes.
Figure 1.
(A) The model protein used to measure the efficiency of membrane integration of designed H segments (Hessa et al., 2005a). If the H-segment forms a transmembrane helix, only the G1 acceptor site for N-linked glycosylation is modified by the lumenal oligosaccharyl transferase (left); if the H-segment does not form a transmembrane helix both the G1 and G2 acceptor sites are modified (right). (B) The “biological” hydrophobicity scale, showing the contribution from each kind of residue to ΔGapp when placed in the middle of a 19-residue-long H-segment (Hessa et al., 2005a). (C) Position-specific contributions to ΔGapp from Arg and Gly residues (Hessa et al., 2005b). ΔGapp values for H segments with the overall composition 1R/6L/12A for the Arg scan and 1G/4L/14A for the Gly scan are plotted against the positions of the Arg and Gly residues in the H segments. Circles indicate the positions of Arg residues in the S4 helix from the KvAP K+ channel (Jiang et al., 2003).
In a first study (Hessa et al., 2005a), we used this approach to measure the contribution to ΔGapp from each of the 20 amino acids when placed in the middle of a 19-residue-long H-segment flanked by “insulating” GGPG….GPGG sequences, Fig. 1 B. The “biological” hydrophobicity scale derived in this way correlates surprisingly well with, e.g., the Wimley-White whole-residue octanol-water partitioning scale (Wimley et al., 1996; White and Wimley, 1999). We have further found that the contribution to ΔGapp from charged residues such as Arg depends strongly on position within the H-segment (Hessa et al., 2005b), Fig. 1 C, possibly explaining how highly charged transmembrane helices like the S4 helix in K+ channel voltage-sensor domains can be stable in a lipid bilayer (see below).
Proline, in itself a nonpolar residue, is very unfavorable when placed in the central and C-terminal part of the H-segment but not when present in the three most N-terminal positions (Hessa et al., 2005b). This pattern is well known in α helices in globular proteins; because of its cyclic side chain, Pro is a helix breaker when placed at the C-terminal but not at the N-terminal end. The obvious implication is that the formation of an α-helix is critical for the membrane insertion of the H-segment. Indeed, some transmembrane α helices have been shown to fold already before inserting into the lipid bilayer (Mingarro et al., 2000; Woolhead et al., 2004; Lu and Deutsch, 2005).
Do Protein–Lipid Interactions Drive Membrane Protein Insertion?
The ΔGapp measurements done so far are fully consistent with the simplest model one can propose for how transmembrane helices are recognized by the ribosome–translocon machinery: that they are somehow allowed to partition into the surrounding lipid bilayer based on the free energy of interaction between the transmembrane segment and the lipid. This would explain the correspondence between the biological hydrophobicity scale and biophysical scales like that of Wimley-White, and it would explain why the positional variations in ΔGapp for residues such as Arg, Trp, Tyr, Phe, and Gly (Hessa et al., 2005a, 2005b) match the statistical distribution of these residues across the membrane in the high-resolution X-ray structures (Ulmschneider et al., 2005). The data at hand thus speak strongly in favor of direct protein–lipid interactions as being the main driving force for the integration of single transmembrane helices, although the translocon may affect the ability of pairs or higher assemblages of transmembrane helices to interact among themselves before partitioning into the bilayer (Meindl-Beinker et al., 2006).
To the extent that this model can be further substantiated, it opens a new experimental window into the detailed physical chemistry of protein–lipid interactions. Where a study of one or a few synthetic peptides interacting with liposomes or other membrane mimetics can take months to complete, H segments can be analyzed en masse in the in vitro transcription–translation system. Suitable adaptations to the experimental setup should also make possible comparative studies on membrane protein integration in different biological membranes (bacterial inner membrane, mitochondrial inner membrane, yeast ER membrane vs. mammalian ER membrane, etc.).
Conversely, if in fact lipid–protein interactions are what matters for natural membrane proteins in vivo, it means that biophysical and computational studies of simplified peptide–lipid systems will be of immediate relevance for understanding the “rules” of membrane protein assembly in the cellular context.
Beyond the “Biological” Hydrophobicity Scale
The technique used to derive the biological hydrophobicity scale avoids many technical problems inherent to more biophysical approaches: no handing of aggregation-prone peptides, no need for the use of spectroscopic probes that may affect the behavior of the peptide, no question of whether a given peptide is transmembrane or not. Of course, there are also shortcomings: the assumption that the ΔGapp values are obtained under equilibrium conditions is hard to test, the “uninserted” state is difficult to characterize (is the H-segment helical or unfolded, is it exposed to solvent, adsorbed to the membrane interface, or perhaps shielded by lumenal chaperones?), the precise way in which an “inserted” H-segment is embedded in the bilayer cannot be determined (which part is helical, is the segment tilted, does it slide vertically relative to the membrane depending on the positions of, e.g., charged residues?), and the lipid composition of the membrane cannot be modified easily, but many of these problems apply also to the biophysical studies.
Although much remains to be done in order to fully understand the results obtained with the Sec61 translocon system, it is notable that the TM1 and TM2 transmembrane helices present in the model protein (see Fig. 1 A) do not seem to affect the results in any significant way, as they can be replaced by a cleavable signal peptide with little effect on the measured ΔGapp values (Meindl-Beinker et al., 2006). Moreover, position-specific contributions to ΔGapp obtained by scanning a single charged or polar residue along an H-segment (e.g., Fig. 1 C) predict ΔGapp values for H segments containing symmetrically disposed pairs of charged residues or even natural transmembrane helices with multiple charged residues within ∼1 kcal/mol (Hessa et al., 2005a,b), suggesting that vertical sliding of the H segments used in the derivation of the “biological” hydrophobicity scale is not a serious problem.
Finally, do these studies tell us anything interesting about voltage-sensor domains? So far, we have focused on the so-called S4 helix in the KvAP voltage sensor (Jiang et al., 2003). This helix contains four Arg residues, yet inserts as a transmembrane helix into the ER membrane with ΔGapp = 0.5 kcal/mol (Hessa et al., 2005b). As seen in Fig. 1 C, the cost for inserting an Arg residue varies strongly with its position in the transmembrane helix, explaining the relatively high membrane insertion propensity of the isolated S4 helix.
This is probably not the whole story, however (see the Perspective by White in this issue, p. 363). Many polar and charged residues, Arg included, have rather long and flexible side chains, making it possible for them to “snorkel” toward the lipid–water interface region. At the same time, lipid molecules located close to a transmembrane helix can adapt to the presence of polar residues, and water molecules can sometimes help solvate polar groups located well within the bilayer plane (Freites et al., 2005; Johansson and Lindahl, 2006; Dorairaj and Allen, 2007); see also the Perspective by MacCallum et al. in this issue (p. 371). One upshot of this dynamic picture of protein–lipid interactions is that ΔGapp profiles such as the one shown in Fig. 1 C most likely do not provide an accurate representation of the free energy profile for moving a charged residue all the way across a membrane (as opposed to inserting it sideways from the translocon as part of a transmembrane helix). Presumably, if a helical peptide is pulled across a lipid bilayer, there is a substantial free energy barrier (not seen in the ΔGapp profile) at the point when a charged residue has to flip its direction of snorkeling from one membrane surface toward the other (Dorairaj and Allen, 2007). Seen from this perspective, one may regard the translocon as a device designed to lower the activation barrier for translocation of polar and charged residues across the membrane by providing an aqueous channel, while at the same time making it possible for consecutive segments of the nascent polypeptide to make “lateral excursions” from the channel in order to test whether the free energy of membrane insertion is favorable or not.
Despite these caveats, it seems likely that the biological hydrophobicity scale is a good measure of the energetics of protein–lipid interactions in the true biological context, and as such will help us define the sequence determinants of membrane protein assembly to a much higher precision than has been possible so far.
Acknowledgments
This work was funded by the grants from the Swedish Research Council, the Marianne and Marcus Wallenberg Foundation, the Swedish Foundation for Strategic Research, and the Swedish Cancer Foundation.
Abbreviations used in this paper: SRP, signal recognition particle.
References
- Alder, N.N., and A.E. Johnson. 2004. Cotranslational membrane protein biogenesis at the endoplasmic reticulum. J. Biol. Chem. 279:22787–22790. [DOI] [PubMed] [Google Scholar]
- Beckmann, R., C.M.T. Spahn, N. Eswar, J. Helmers, P.A. Penczek, A. Sali, J. Frank, and G. Blobel. 2001. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell. 107:361–372. [DOI] [PubMed] [Google Scholar]
- Dorairaj, S., and T.W. Allen. 2007. On the thermodynamic stability of a charged arginine side chain in a transmembrane helix. Proc. Natl. Acad. Sci. USA. In press. [DOI] [PMC free article] [PubMed]
- Freites, J.A., D.J. Tobias, G. von Heijne, and S.H. White. 2005. Interface connections of a transmembrane voltage sensor. Proc. Natl. Acad. Sci. USA. 102:15059–15064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessa, T., H. Kim, K. Bihlmaier, C. Lundin, J. Boekel, H. Andersson, I. Nilsson, S.H. White, and G. von Heijne. 2005. a. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 433:377–381. [DOI] [PubMed] [Google Scholar]
- Hessa, T., S.H. White, and G. von Heijne. 2005. b. Membrane insertion of a potassium channel voltage sensor. Science. 307:1427. [DOI] [PubMed] [Google Scholar]
- Jiang, Y., A. Lee, J. Chen, V. Ruta, M. Cadene, B.T. Chait, and R. MacKinnon. 2003. X-ray structure of a voltage-dependent K+ channel. Nature. 423:33–41. [DOI] [PubMed] [Google Scholar]
- Johansson, A.C., and E. Lindahl. 2006. Amino-acid solvation structure in transmembrane helices from molecular dynamics simulations. Biophys. J. 91:4450–4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, J., and C. Deutsch. 2005. Secondary structure formation of a transmembrane segment in Kv channels. Biochemistry. 44:8230–8243. [DOI] [PubMed] [Google Scholar]
- Meindl-Beinker, N.M., C. Lundin, I. Nilsson, S.H. White, and G. von Heijne. 2006. Asn- and Asp-mediated interactions between transmembrane helices during translocon-mediated membrane protein assembly. EMBO Rep. 7:1111–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingarro, I., I. Nilsson, P. Whitley, and G. von Heijne. 2000. Different conformations of nascent polypeptides during translocation across the ER membrane. BMC Cell Biol. 1:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra, K., C. Schaffitzel, T. Shaikh, F. Tama, S. Jenni, C.L. Brooks III, N. Ban, and J. Frank. 2005. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature. 438:318–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sääf, A., E. Wallin, and G. von Heijne. 1998. Stop-transfer function of pseudo-random amino acid segments during translocation across prokaryotic and eukaryotic membranes. Eur. J. Biochem. 251:821–829. [DOI] [PubMed] [Google Scholar]
- Ulmschneider, M.B., M.S. Sansom, and A. Di Nola. 2005. Properties of integral membrane protein structures: derivation of an implicit membrane potential. Proteins. 59:252–265. [DOI] [PubMed] [Google Scholar]
- van den Berg, B., W.M. Clemons, I. Collinson, Y. Modis, E. Hartmann, S.C. Harrison, and T.A. Rapoport. 2004. X-ray structure of a protein-conducting channel. Nature. 427:36–44. [DOI] [PubMed] [Google Scholar]
- White, S.H. 2004. The progress of membrane protein structure determination. Protein Sci. 13:1948–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, S.H., and W.C. Wimley. 1999. Membrane protein folding and stability: physical principles. Annu. Rev. Biophys. Biomol. Struct. 28:319–365. [DOI] [PubMed] [Google Scholar]
- Wimley, W.C., T.P. Creamer, and S.H. White. 1996. Solvation energies of amino acid sidechains and backbone in a family of host-guest pentapeptides. Biochemistry. 35:5109–5124. [DOI] [PubMed] [Google Scholar]
- Woolhead, C.A., P.J. McCormick, and A.E. Johnson. 2004. Nascent membrane and secretory proteins differ in FRET-detected folding. Cell. 116:725–736. [DOI] [PubMed] [Google Scholar]