Small RNAs (sRNAs), often called noncoding RNAs, are widespread in all organisms and are used to control diverse cellular processes. A large class of sRNAs is involved in the regulation of gene expression primarily at posttranscriptional levels through base-pairing. MicroRNAs and short interfering RNAs represent this class of sRNAs in eukaryotes, whereas Hfq-binding sRNAs are the major base-pairing sRNAs in bacteria. Since the serendipitous discovery of a base-pairing RNA in Escherichia coli (1), a number of chromosomally encoded base-pairing sRNAs have been identified in bacteria. Although the functions of many of them remain to be elucidated, an emerging view is that these sRNAs act as important players in regulatory cascades consisting of diverse physiological processes. In E. coli, most base-pairing sRNAs are induced under specific stress conditions, and regulate, mostly negatively, translation and stability of target mRNAs through an imperfect base-pairing depending on the RNA chaperone Hfq (2). The changes of expression of target mRNAs contribute to the stress response. It has been assumed that Hfq-binding sRNAs do not encode proteins and execute their functions only through base-pairing mechanisms, referred to as riboregulation. In this issue of PNAS, Wadler and Vanderpool (3) demonstrate that this view must be changed in some instances. The authors make the exciting discovery that an Hfq-binding sRNA of E. coli acts not only by base-pairing but also by serving as an mRNA template for a small functional protein to deal with the same metabolic stress.
In E. coli, external glucose is transported into the cells coupled with phosphorylation resulting in glucose-6-phosphate (G6P). The major glucose transporter, IICBGlc, is encoded by ptsG (Fig. 1). When the glycolytic flux is blocked, for example by a mutation in pgi encoding phosphoglucose isomerase, the intracellular level of G6P greatly increases (4). The accumulation of G6P represents a metabolic stress because it is somehow toxic for cells (5). An analogous situation occurs when the wild-type cells are exposed to a nonmetabolizable glucose analog α-methyl-glucoside (αMG). Until recently, little was known about how cells deal with the metabolic stress caused by intracellular accumulation of phosphosugars. Several years ago, it was found that the glucose–phosphate stress causes a striking reduction of IICBGlc expression and an RNase E-dependent rapid degradation of ptsG mRNA (6). The physiological relevance of this down-regulation of ptsG in response to glucose–phosphate stress would be to avoid further accumulation of glucose phosphates.
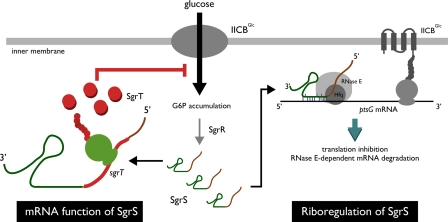
Fig. 1.
Dual function of SgrS under glucose phosphate stress. (Middle) Glucose is transported and phosphorylated into the cell by IICBGlc. When G6P accumulates abnormally, the synthesis of SgrS is induced, depending on SgrR. (Right) SgrS forms a ribonucleoprotein complex associating with Hfq and RNase E to act on the ptsG mRNA encoding IICBGlc through base-pairing, resulting in translational inhibition and RNase E-dependent rapid degradation of the message (17). The physiological role of SgrS-mediated riboregulation of ptsG mRNA is to limit accumulation of toxic sugar phosphates by stopping new synthesis of IICBGlc. (Left) As shown by Wadler and Vanderpool (3) in this issue of PNAS, SgrS acts also as an mRNA template for a small functional protein SgrT to prevent glucose uptake, presumably by inhibiting the transport activity of IICBGlc. The SgrT function should be useful to rapidly prevent glucose uptake by inhibiting preexisting IICBGlc, whereas the riboregulation through base-pairing would be important for adaptation to prolonged stress. It is not known whether one SgrS molecule is involved in both base-pairing and SgrT production or different SgrS molecules exert the two functions separately.
Vanderpool and Gottesman (7) found that overproduction of an Hfq-binding sRNA of unknown function, now called SgrS (sugar transport-related sRNA), inhibits cell growth on glucose, raising the possibility that SgrS is involved in the down-regulation of ptsG. Indeed, they demonstrated that the synthesis of SgrS is induced in response to glucose–phosphate stress and noticed that an ≈30-nt region within the 3′ portion of SgrS is partially complementary to the translation initiation region of ptsG mRNA. These observations led them to propose that SgrS down-regulates ptsG mRNA through a base-pairing mechanism (Fig. 1 Right). Consistent with this model, ptsG mRNA remains stable during the stress in cells lacking SgrS. Another gene, sgrR, encoding a putative transcription factor, is required for the induction of SgrS under the stress condition. Further studies have uncovered several intriguing features regarding the mechanism of SgrS action (Fig. 1 Right). Hfq binds to RNase E, the major endoribonuclease in E. coli, and SgrS RNA associates with RNase E through Hfq to form a specific ribonucleoprotein complex, which is apparently required for the rapid degradation of ptsG mRNA (8). Nevertheless, translational inhibition rather than mRNA degradation is primarily responsible for gene silencing (9). Membrane localization of the target mRNA facilitates the action of SgrS presumably by affecting competition between SgrS and ribosomes (10). Hfq stimulates the SgrS-ptsG base-pairing by accelerating the rate of duplex formation (11). A short region consisting of six contiguous base-pairs around the ribosome binding site is crucial for the action of SgrS on ptsG mRNA (11).
The physiological importance of SgrS is clear because cells lacking the sgrS gene are severely inhibited for growth, whereas wild-type cells are only transiently inhibited under the glucose–phosphate stress condition (7, 9). An obvious role of SgrS is to inhibit synthesis of IICBGlc through riboregulation, leading to reduced glucose uptake and blocking further accumulation of G6P. However, stopping new synthesis of IICBGlc alone is apparently not enough to prevent further accumulation of G6P under stress conditions because preexisting IICBGlc would continue to take up glucose. How do cells deal with this potential problem? Now, Wadler and Vanderpool (3) show that SgrS itself acts as an mRNA template to make a small protein, an inhibitor of the glucose transporter.
SgrS is ≈220 nt in length, which is longer than most other Hfq-binding sRNAs. The region required for base-pairing with ptsG mRNA is located in the 3′ portion of SgrS (7, 11). This leads to a question: what is the role of the 5′ portion of SgrS? Wadler and Vanderpool (3) found an ORF for a putative polypeptide of 43 aa designated SgrT upstream of the base-pairing region within the 5′ portion of SgrS (Fig. 1 Left). The sgrT gene is fairly conserved in many bacterial species, suggesting the functional importance of the SgrT protein. To examine the role of SgrT, several modified sgrS genes under the control of an IPTG-inducible promoter were constructed on plasmids; these expressed only the base-pairing, only the SgrT ORF, both, or neither. Either base-pairing or SgrT was sufficient to protect cells from the stress of αMG treatment, and either was sufficient to block growth on glucose, suggesting that they have redundant functions in helping the cell deal with this stress. Indeed, the authors showed that either base-pairing or SgrT was able to block glucose uptake efficiently. SgrT had no effect on either ptsG mRNA or IICBGlc protein levels, suggesting that it acts independently of base-pairing, synthesis, or degradation of the mRNA and protein. Therefore, the authors propose that SgrT inhibits the activity of IICBGlc presumably by “plugging” the transport channel or inhibiting IICBGlc phosphorylation based on protein–protein interactions (Fig. 1 Left). Thus, SgrT may help block the action of preexisting protein, whereas the base-pairing blocks new synthesis.
Many “noncoding” RNAs await characterization for potential hidden functions.
The discovery of SgrT has settled a question of how cells prevent glucose uptake through preexisting IICBGlc. SgrT may prevent glucose uptake immediately by inhibiting IICBGlc activity when it is translated. On the other hand, the base-pairing blocks new synthesis of IICBGlc, which would be particularly important for adaptation to prolonged stress. If this is the case, one can expect that the riboregulation and SgrT function cooperate to deal with the same glucose–phosphate stress through distinct mechanisms. However, the authors showed that either of two activities of SgrS, the base-pairing or SgrT, is sufficient for SgrS function regarding glucose uptake inhibition and therefore growth phenotypes when SgrS derivatives were artificially overexpressed. Why are two activities functionally redundant rather than cooperative? Is this also true in its natural context? The SgrT protein seems to be expressed at a very low level under the stress in wild-type cells. Is this low amount of SgrT sufficient to mediate the rapid response to the stress? How does SgrT inhibit the activity of glucose PTS? Does the same SgrS molecule that is used for base-pairing also get translated for SgrT production, or are different SgrS molecules used? Some of these important questions will be harder to answer than others, but we expect they will be worked out in the near future.
There are two other examples for bifunctional bacterial sRNAs that also serve as an mRNA. One is RNAIII of Staphylococcus aureus that functions by base-pairing with several mRNA targets related to bacterial virulence and also encodes a small peptide (12, 13). The riboregulation mediated by RNAIII does not require Hfq, and there is no evidence that the small peptide acts to affect the proteins regulated by the base-pairing. Apart from base-pairing sRNAs, another well known example is tmRNA, found in all eubacteria, that acts as an alanyl tRNA and an mRNA (14, 15). The small peptide encoded by tmRNA is used to mark incomplete nascent polypeptides during an unusual translation process called trans-translation. Thus, SgrS is the first example of an Hfq-binding bifunctional sRNA encoding a functional small protein. The most important lesson to be learned from the work of Wadler and Vanderpool (3) is that sRNAs may posses the ability to encode small functional polypeptides in some cases. It is rather natural to assume that more examples for Hfq-binding bifunctional sRNAs will be discovered. So far, no example for eukaryotic sRNAs encoding a small protein is known. It should be noted, however, that a poly(A)-containing RNA, annotated as a noncoding RNA, has been shown to encode a small peptide involved in the regulation in cell morphogenesis in Drosophila (16). Many “noncoding” RNAs await characterization for potential hidden functions.
Footnotes
The authors declare no conflict of interest.
See companion article on page 20454.
References
- 1.Mizuno T, Chou MY, Inouye M. Proc Natl Acad Sci USA. 1984;81:1966–1970. doi: 10.1073/pnas.81.7.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Storz G, Gottesman S. Versatile Roles of Small RNA Regulators in Bacteria. Plainview, NY: Cold Spring Harbor Lab Press; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wadler CS, Vanderpool CK. Proc Natl Acad Sci USA. 2007;104:20454–20459. doi: 10.1073/pnas.0708102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morita T, El-Kazzaz W, Tanaka Y, Inada T, Aiba H. J Biol Chem. 2003;278:15608–15614. doi: 10.1074/jbc.M300177200. [DOI] [PubMed] [Google Scholar]
- 5.Lee AT, Cerami A. Proc Natl Acad Sci USA. 1987;84:8311–8314. doi: 10.1073/pnas.84.23.8311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kimata K, Tanaka Y, Inada T, Aiba H. EMBO J. 2001;20:3587–3595. doi: 10.1093/emboj/20.13.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vanderpool CK, Gottesman S. Mol Microbiol. 2004;54:1076–1089. doi: 10.1111/j.1365-2958.2004.04348.x. [DOI] [PubMed] [Google Scholar]
- 8.Morita T, Maki K, Aiba H. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morita T, Mochizuki Y, Aiba H. Proc Natl Acad Sci USA. 2006;103:4858–4863. doi: 10.1073/pnas.0509638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawamoto H, Morita T, Shimizu A, Inada T, Aiba H. Genes Dev. 2005;19:328–338. doi: 10.1101/gad.1270605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawamoto H, Koide Y, Morita T, Aiba H. Mol Microbiol. 2006;61:1013–1022. doi: 10.1111/j.1365-2958.2006.05288.x. [DOI] [PubMed] [Google Scholar]
- 12.Huntzinger E, Boisset S, Saveanu C, Benito Y, Geissmann T, Namane A, Lina G, Etienne J, Ehresmann B, Ehresmann C, et al. EMBO J. 2005;24:824–835. doi: 10.1038/sj.emboj.7600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, et al. Genes Dev. 2007;21:1353–1366. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keiler KC, Waller PR, Sauer RT. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 15.Keiler KC. Curr Opin Microbiol. 2007;10:169–175. doi: 10.1016/j.mib.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 16.Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S, Kageyama Y. Nat Cell Biol. 2007;9:660–665. doi: 10.1038/ncb1595. [DOI] [PubMed] [Google Scholar]
- 17.Aiba H. Curr Opin Microbiol. 2007;10:134–139. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]