Abstract
We describe a convenient method for distinguishing functional from nonfunctional or deleterious mutations in homologous genes. High fidelity in vitro gene recombination (“DNA shuffling”) coupled with sequence analysis of a small sampling of the shuffled library exhibiting the evolved behavior allows identification of those mutations responsible for the behavior in a background of neutral and deleterious mutations. Functional mutations are expected to occur in 100% of the sequenced screened sample; neutral mutations are found in 50% on average, and deleterious mutations do not appear at all. When used to analyze 10 mutations in a laboratory-evolved gene encoding a thermostable subtilisin E, this method rapidly identified the two responsible for the observed protease thermostability; the remaining eight were neutral with respect to thermostability, within the precision of the screening assay. A similar approach, coupled with selection for growth and survival of the host organism, could be used to distinguish adaptive from neutral mutations.
Keywords: adaptive evolution, neutral mutations, DNA shuffling, in vitro evolution
A fundamental problem in the study of evolutionarily related genes is to distinguish those mutations responsible for phenotypic differences from a background of neutral mutations that have little or no effect on function. In nature, adaptive changes may represent only a small fraction of all evolutionary events (1, 2). Some fraction of these may lead to functional differences measured in vitro. It is difficult to identify with certainty specific adaptive mutations; even mutations responsible for specific functional differences among proteins can evade identification when multiple nonfunctional mutations are present (3). Thus, the use of sequence comparisons is of limited utility in identifying the molecular mechanisms underlying differences in properties such as thermostability exhibited by proteins encoded by related genes.
This problem exists for sequences evolved in vitro as well. Although in vitro evolution can lead to the development of useful new protein functions, the responsible mutations almost always occur in a background of mutations that are neutral or even deleterious to the behavior(s) of interest. To derive key information on structure–activity relationships from these and nature’s own experiments in molecular evolution, a convenient method for distinguishing functional from nonfunctional and/or deleterious mutations is needed. Site-directed mutagenesis requires the construction of multiple variants with different combinations of mutations and is far too laborious when many mutations are present. Here we demonstrate how a single experiment involving the random recombination of homologous sequences followed by screening for the altered behavior can be used to identify functional mutations. The experiment is based on the in vitro “DNA shuffling” method developed by Stemmer (4), with modifications to dramatically reduce the associated point mutagenesis rate (5). DNA shuffling of homologous genes creates a library of genes containing all possible combinations of mutations. As shown here, functional mutations are identified upon sequencing a set of the recombined genes that exhibit the evolved property. This approach, coupled with selection rather than screening, could be used to distinguish adaptive (i.e., those affecting the growth and survival of the organism) from neutral mutations.
MATERIALS AND METHODS
Restriction enzymes were purchased from Boehringer Mannheim. Succinyl-Ala-Ala-Pro-Phe-p-nitroanilide was from Sigma. Bacillus subtilis strain DB428 and Bacillus cloning vector pKWZ containing the subtilisin E gene were kindly provided by Dr. R. Doi of University of California, Davis, CA.
DNA Shuffling and Screening for Thermostability.
High fidelity DNA shuffling of wild-type subtilisin E gene and 1E2A is described elsewhere (5). The PCR-amplified, reassembled product was purified by Wizard PCR prep kit (Promega), digested with BamHI and NdeI, and electrophoresed in a 0.8% agarose gel. The 986-bp product was cut from the gel and purified by QIAEX II gel extraction kit (Qiagen, Chatsworth, CA). Products were ligated with vector generated by BamHI–NdeI digestion of the pBE3 Escherichia coli–B. subtilis shuttle vector (Fig. 1). This gene library was amplified in E. coli HB101 and transferred into B. subtilis DB428-competent cells as described (6); 768 clones were picked with sterile toothpicks and grown in SG medium supplemented with 50 μg/ml kanamycin at 37°C for 24 h in eight 96-well plates. The cells were spun down, and samples of the supernatants were examined in the thermostability assay. Three replica 96-well assay plates were duplicated for each growth plate, with each well containing 5 μl of supernatant. Enzyme activity was measured as described (6) in 96-well plates using a Thermomax microplate reader (Molecular Devices). Activity measured at room temperature was used to calculate the fraction of active clones. Clones with activity <10% of that of wild type were scored as inactive. Initial activity (Ai) was measured on one assay plate after incubation at 65°C for 10 min by adding 100 μl of prewarmed (37°C) assay solution [0.2 mM succinyl-Ala-Ala-Pro-Phe-p-nitroanilide (suc-AAPF-pNA)/100 mM Tris⋅HCl, pH 8.0/10 mM CaCl2] into each well. Residual activity (Ar) was measured after 40 min of incubation.
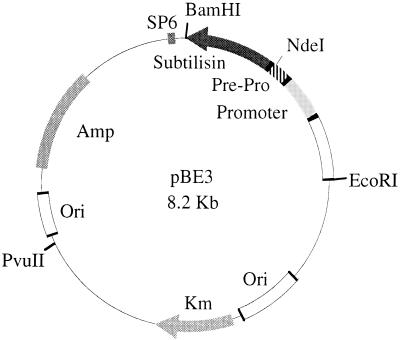
Figure 1.
E. coli/B. subtilis shuttle vector pBE3 containing the β-lactamase (Ampr) gene and replicon from pGEM3 for growth in E. coli and the kanamycin nucleotidyl transferase (Kmr) gene and replicon from pUB110 for growth in B. subtilis. The subtilisin E gene including its natural promoter was subcloned from plasmid pKWZ by BamHI and EcoRI restriction sites.
Sequence Analysis.
Genes were individually purified from B. subtilis DB428 using a QIAprep spin plasmid miniprep kit (Qiagen) with the modifications that 2 mg/ml lysozyme was added to P1 buffer, and the cells were incubated for 5 min at 37°C, retransformed into competent E. coli HB101, and then purified again using QIAprep spin plasmid miniprep kit to obtain sequencing quality DNA. Sequencing was done on an Applied Biosystems 373 DNA Sequencing System using the Dye Terminator Cycle Sequencing kit (Perkin–Elmer).
Construction of Subtilisin E Variants by Site-Directed Mutagenesis.
Site-directed mutagenesis was carried out using an Altered Sites II in vitro mutagenesis kit (Promega). The wild-type gene was subcloned into pAlter-Ex1 vector with BamHI and EcoRI. For each substitution (V93I, N109S, N181D, N218S), an oligonucleotide containing the desired mutation was used as the mutagenic primer. Mutants were screened by direct partial sequencing. Desired clones were subcloned back to the pBE3 shuttle vector digested with NdeI and BamHI. All mutations were confirmed by DNA sequencing.
Enzyme Purification and Thermal Inactivation Assay.
All of the subtilisin E variants were purified as described (6). Purified variants (≈10 μM) were dialyzed in 10 mM Tris⋅HCl/1 mM CaCl2 (pH 8.0) at 4°C overnight. The samples were incubated at 65°C on a thermocycler. At various time intervals, 10-μl aliquots were added to 0.99 ml of activity assay solution (0.2 mM suc-AAPF-pNA/100 mM Tris⋅HCl/10 mM CaCl2, pH 8.0, 37°C). Thermal inactivation at 65°C of all of the subtilisin E variants appears to obey first-order kinetics; t1/2 is the half-life of enzyme activity at 65°C.
Specific Activity.
Specific activity (unit/mg) was determined as the ratio of enzyme activity to protein concentration, under the same conditions as in thermal inactivation assay (0.2 mM suc-AAPF-pNA/100 mM Tris⋅HCl/10 mM CaCl2, pH 8.0, 37°C). One unit will hydrolyze suc-AAPF-pNA to produce the color equivalent to 1.0 μmol of p-nitroanilide per min at pH 8.0, 37°C.
Differential Scanning Calorimetry.
Melting temperatures (Tm) were determined by differential scanning calorimetry on a MicroCal MS1 calorimeter (Microcal, Amherst, MA). Experiments were carried out in 10 mM Tris⋅HCl/1 mM CaCl2/1 mM phenylmethylsulfonylfluoride (an inhibitor of subtilisin E), pH 8.0. The temperature was increased at a rate of 1°C/min from 20°C to 90°C. The protein concentration (≈30 μM) was determined by absorbance at 280 nm (ɛ = 35886 M−1⋅cm−1), and the sample size was 1.769 ml.
RESULTS AND DISCUSSION
Thermostable subtilisin E variant 1E2A was obtained by in vitro evolution of the wild-type enzyme (5, 6). At 65°C, its rate of thermoinactivation is about 8-fold slower than that of the wild-type protease. The mature gene encoding 1E2A differs from the wild-type sequence at 10 base positions; six mutations are synonymous with respect to the amino acid sequence, and four lead to amino acid changes (Table 1). All four nonsynonymous mutations exist in other naturally occurring subtilisins. To determine which mutations are responsible for the increased thermostability of the enzyme, we randomly recombined the 1E2A and wild-type subtilisin E genes to create a library of sequences containing all possible combinations of the mutations. This library was then expressed and screened to identify thermostable enzymes. In theory, the mutations responsible for the increased thermostability of the evolved enzyme should be present in 100% of the sequences coding for enzymes whose thermostability equals that of 1E2A subtilisin E. If deleterious mutations were present in the original evolved sequence, as can often happen when multiple mutations are accumulated in a single generation of directed evolution (7), then they would be removed in the recombined population that passed the screen for the evolved function. Thus, deleterious mutations would be present at 0% frequency in the sequenced genes. Mutations that have no effect on function should be present in roughly 50% of the screened sequences, if equimolar amounts of the parental genes are shuffled.
Table 1.
DNA and amino acid substitutions in thermostable 1E2A subtilisin E (mature gene)
| Base | Base substitution | Position in codon | Amino acid | Amino acid substitution |
|---|---|---|---|---|
| 484 | A → G | 3 | 10 | synonymous |
| 520 | A → T | 3 | 22 | synonymous |
| 731 | G → A | 1 | 93 | Val → Ile |
| 745 | T → C | 3 | 97 | synonymous |
| 780 | A → G | 2 | 109 | Asn → Ser |
| 995 | A → G | 1 | 181 | Asn → Asp |
| 1107 | A → G | 2 | 218 | Asn → Ser |
| 1141 | A → T | 3 | 229 | synonymous |
| 1153 | A → G | 3 | 233 | synonymous |
| 1189 | A → G | 3 | 245 | synonymous |
High Fidelity DNA Shuffling.
As described (5), an equimolar mixture of the ≈1-kb wild-type and 1E2A genes was fragmented with DNase I, and the column-purified 20- to 50-bp fragments were reassembled (initially without primer) to a single PCR product of the correct size. The results of gene fragmentation, reassembly, and amplification are shown in Fig. 2. The overall rate of point mutagenesis associated with the high fidelity DNA shuffling of these genes is only 0.05% (5). After DNA shuffling, the gene library was subcloned and amplified in E. coli and then transferred into B. subtilis-competent cells for expression and screening. This was facilitated by an E. coli/B. subtilis shuttle vector, pBE3 (Fig. 1). This vector contains two sets of antibiotic resistance genes and replicons; one is functional in E. coli and the other in B. subtilis (although the kanamycin resistance gene is also expressed at low levels in E. coli). The transformation efficiency by ligated vectors is 3 × 105 cfu/μg for Ca2+-prepared E. coli competent cells, and the transformation efficiency by plasmid DNA is 2 × 104 cfu/μg for B. subtilis DB428-competent cells. The direct cloning of recombined DNA into B. subtilis competent cells occurs at very low efficiency (less than a few hundred transformants per μg of DNA), and use of this shuttle vector greatly enhances the size of the recombined and/or randomly mutated gene libraries that can be created in B. subtilis.
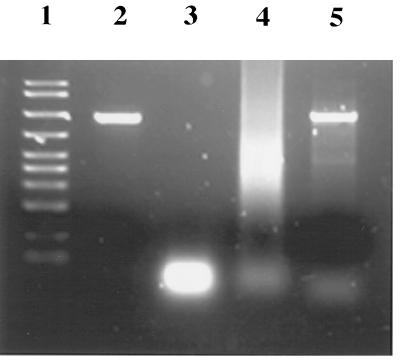
Figure 2.
DNA agarose gel (2%) showing process of high fidelity DNA shuffling of 1E2A and wild-type (wt) subtilisin E genes (5). Lanes: 1, AmpliSize DNA Size standards (Bio-Rad), from top to bottom: 2000, 1500, 1000, 700, 500, 400, 300, 200, 100, and 50 bp; 2, 1:1 mixture of 986-bp fragments from wt and 1E2A before DNase I digestion; 3, 1:1 DNA mixture of wt and 1E2A after 2 min of DNase I digestion in the presence of Mn2+; 4, fragment reassembly with Pfu polymerase; and 5, PCR amplification of the reassembly product with Taq/Pfu (1:1). Primers 5′-CCGAG CGTTG CATAT GTGGA AG-3′ (underlined sequence is NdeI restriction site) and 5′-CGACT CTAGA GGATC CGATT C-3′ (underlined sequence is BamHI restriction site) were used.
Sequence Analysis.
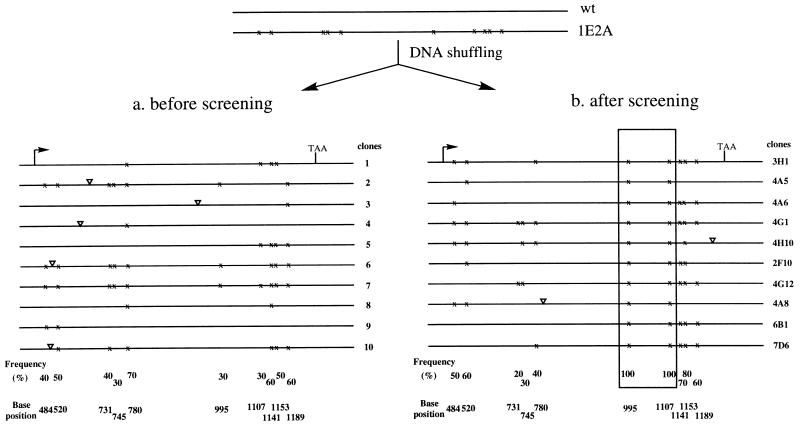
As a control, 10 unscreened clones from the recombined gene library were selected at random and sequenced (5). The results are summarized in Fig. 3a. All except clone 7 result from different recombination events. (Clone 7 is the intact 1E2A parent sequence.) The frequency of occurrence of a particular point mutation from parent 1E2A in the shuffled genes ranged from 30 to 70%, fluctuating around the expected value of 50%. All 10 mutations can be recombined, even those that are only 12 bp apart. There is clear linkage, however, between such closely spaced mutations.
Figure 3.
Sequence analysis of randomly recombined gene libraries before (a) and after (b) screening for protease thermostability. Lines represent 986 bp of subtilisin E gene including 45 nt of its prosequence, the entire mature sequence, and 113 nt after the stop codon. Crosses indicate positions of mutations from 1E2A, and triangles indicate positions of new point mutations introduced during the DNA shuffling procedure.
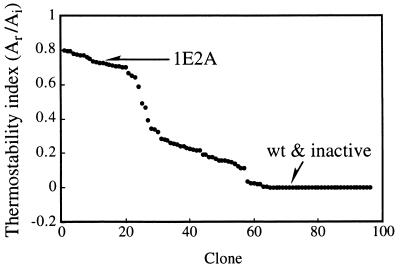
We then assayed the rates of subtilisin thermoinactivation at 65°C for 768 clones picked from SG agar plates with 50 μg/ml kanamycin and grown in eight 96-well plates. Initial activity (Ai) was measured on the culture supernatants in each well after incubation at 65°C for 10 min. Residual activity (Ar) was measured after 40 min of incubation. The normalized residual activity (Ar/Ai) was used as an index of thermostability. Thermostabilities measured on a typical 96-well plate are shown in Fig. 4, plotted in descending order. Approximately 23% of the clones exhibited thermostability comparable to 1E2A, which indicates immediately that only two mutations [(1/2)2 = 25%] are responsible for the increased thermostability.† Twenty clones exhibiting the highest thermostability were selected, and their kinetics of thermoinactivation were verified in a second assay on the culture supernatants (assay used for purified enzymes as described in Materials and Methods). No false-positives were found. Genes from the 10 most thermostable were sequenced (Fig. 3b). Only two new point mutations were found among these 10 recombined, screened genes, as compared with five in the unscreened population (Fig. 3a). Most point mutations are deleterious to thermostability (data not shown). The lower rate of point mutations found in the screened population reflects the “cleansing” effect of the screen (8).
Figure 4.
Results of screening a typical 96-well plate for residual activity after incubation at 65°C for 10 (Ai) and 40 min (Ar). Ar/Ai was used as an index of the enzyme’s thermostability. Data are sorted and plotted in descending order.
The two nonsynonymous mutations leading to amino acid substitutions N181D and N218S were found in all 10 of the recombined clones exhibiting high thermostability. The remaining eight mutations occurred at frequencies of 20–80%, very similar to the frequencies observed in the unscreened control sample (Fig. 4a). Thus, we can conclude that N181D and N218S are functional mutations and that the rest are neutral with respect to thermostability. In addition, we can conclude that none of the 10 mutations is deleterious, at least within the precision of the assay method, which is sensitive to changes on the order of 15% in deactivation rate.
Stability and Activity of Specific Subtilisin E Variants.
To verify this result, all four nonsynonymous single mutants and the N181D+ N218S double mutant were constructed by site-directed mutagenesis and characterized with respect to their activities and thermostabilities (Table 2). 1E2A and double mutant N181D+N218S have similar half-lives of thermoinactivation at 65°C and similar melting temperatures, Tm. The half-lives at 65°C of single mutants N181D and N218S are ≈3- and 2-fold greater than that of wild-type subtilisin E, respectively, and their Tms are 3.7 and 3.2°C higher. Thus, both functional mutations identified by random recombination also confer thermostability in the wild-type background. Mutation N218S was found previously in a thermostable variant of subtilisin BPN′ (9). N181D and N218S also are found in thermostable subtilisin E homologs thermitase (10), proteinase K (10), aerolysin (11), and Ak1 proteinase (12). In contrast, the half-lives at 65°C of the single mutants V93I and N109S are very similar to that of wild-type, as are their Tm values. None of the four amino acid substitutions decreased the specific activity of the enzyme; N218S increased both specific activity and stability (Table 2).
Table 2.
Stabilities and activities of purified subtilisin E variants
| Variant | t1/2 at 65°C*, min | Tm†, °C | Specific activity, unit/mg‡ |
|---|---|---|---|
| Wild type | 5.1 ± 0.2 | 68.1 | 17.2 ± 0.1 |
| 1E2A | 42.8 ± 0.1 | 74.4 | 33.7 ± 0.7 |
| V93I | 5.0 ± 0.1 | 68.1 | 21.6 ± 0.2 |
| N109S | 5.2 ± 0.1 | 68.2 | 16.1 ± 0.4 |
| N181D | 16.5 ± 0.6 | 71.8 | 18.0 ± 0.6 |
| N218S | 10.9 ± 0.1 | 71.3 | 38.6 ± 0.6 |
| N218S + N181D | 49.9 ± 0.8 | 74.6 | 38.4 ± 0.1 |
Half-life of thermoinactivation, pH 8.0, 10 mM CaCl2.
Melting temperature, as measured by differential scanning calorimetry, pH 8.0, 1 mM CaCl2.
Specific activity toward suc-AAPF-pNA at pH 8.0.
Although thermophilic organisms in nature have evolved extremely stable enzymes, high stability has often come at the cost of specific activity at the lower temperature. This trade-off, however, may reflect evolutionary drift or even certain requirements for function within an adaptive biological network. Thus, it may not necessarily hold true for in vitro evolution, where the enzyme is decoupled from its natural function. Mutations that increase thermostability without decreasing specific activity are not extremely rare. Furthermore, it should be possible to combine evolved properties of thermostability and enhanced activity.
The method outlined above can be used to identify functional mutations in far more complicated systems than the laboratory-evolved thermostable subtilisin E used here for demonstration. For example, thermostable and nonthermostable members of a protein family often differ not at four but at dozens of amino acids. To identify those amino acid substitutions conferring thermostability using site-directed mutagenesis would require construction and characterization of an impractically large number of variants. For all practical purposes, functional mutations can be identified by screening a randomly recombined gene library when the number of functional mutations is on the order of 10 or less in a background of any number of neutral (or deleterious) mutations (provided there is sufficient homology between the genes for the in vitro recombination to succeed). With 10 functional mutations, the frequency of the thermostable phenotype would be (1/2)10 ≈ 0.1%. Thus, ≈10 thermostable clones could be identified by screening 10,000 clones, which is quite feasible using a simple 96-well plate assay. It is likely that an even larger number of functional mutations could be distinguished by coupling the evolved phenotype to a functional selection rather than a screen.
The experiment we have described differs in several important aspects from simple back-crossing with an excess of a parental sequence (or wild type) by DNA shuffling (4), which will effectively flush out both neutral and deleterious mutations because of the statistical preference for incorporating the parental sequence in the recombined genes (J. C. Moore, H. M. Jin, O. Kuchner, and F.H.A., unpublished work). Using an excess of parental sequence dramatically decreases the frequency of clones exhibiting the evolved property and therefore greatly increases the screening requirement. For example, in a library created by back-crossing a gene containing five functional mutations with a 10-fold excess of the wild-type sequence, the frequency of the evolved phenotype would be only (1/11)5, or 6.2 × 10−6. Furthermore, the relatively high rate of mutagenesis in the original Stemmer protocol, 0.7% (corresponding to ≈7 new mutations per gene) would mask the relationship between evolved phenotype and functional mutations that forms the basis of this experiment. Finally, this experiment is capable of distinguishing the deleterious mutations (0% expected frequency) from those that are neutral (50% frequency). Such a distinction would not be possible if the Stemmer method were used to back-cross with excess wild-type sequence.
Acknowledgments
We thank Prof. Steven Benner (University of Florida) for helpful discussions. This work was supported by the U. S. Department of Energy’s program in Biological and Chemical Technologies Research within the Office of Industrial Technologies, Energy Efficiency and Renewables.
Footnotes
The probability that any given mutation will appear in the randomly recombined population is 1/2. Thus, the probability that N-specific (functional) mutations will appear together in a sequence is (1/2)N.
References
- 1.Stewart C B, Schilling J W, Wilson A C. Nature (London) 1987;330:401–404. doi: 10.1038/330401a0. [DOI] [PubMed] [Google Scholar]
- 2.Perutz M F. Mol Biol Evol. 1983;1:1–28. doi: 10.1093/oxfordjournals.molbev.a040299. [DOI] [PubMed] [Google Scholar]
- 3.Benner S A. Chem Rev. 1989;89:789–806. [Google Scholar]
- 4.Stemmer W P C. Nature (London) 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 5.Zhao H, Arnold F H. Nucleic Acids Res. 1997;25:1307–1308. doi: 10.1093/nar/25.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen K, Arnold F H. Bio/Technology. 1991;9:1073–1077. doi: 10.1038/nbt1191-1073. [DOI] [PubMed] [Google Scholar]
- 7.Moore J C, Arnold F H. Nat Biotechnol. 1996;14:458–467. doi: 10.1038/nbt0496-458. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki M, Christians F C, Kim B, Skandalis A, Black M E, Loeb L A. Mol Diversity. 1996;2:111–118. doi: 10.1007/BF01718708. [DOI] [PubMed] [Google Scholar]
- 9.Bryan P N, Rollence M L, Pantoliano M W, Wood J, Finzel B C, Gilliland G L, Howard A J, Poulos T L. Proteins Struct Funct Genet. 1986;1:326–334. doi: 10.1002/prot.340010406. [DOI] [PubMed] [Google Scholar]
- 10.Siezen R J, Devos W M, Leunissen J A M, Dijkstra B W. Protein Eng. 1991;4:719–737. doi: 10.1093/protein/4.7.719. [DOI] [PubMed] [Google Scholar]
- 11.Volkl P, Markiewicz P, Stetter K O, Miller J H. Protein Sci. 1994;3:1329–1340. doi: 10.1002/pro.5560030819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maciver B, Mchale R H, Saul D J, Bergquist P L. Appl Environ Microbiol. 1994;60:3981–3988. doi: 10.1128/aem.60.11.3981-3988.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]