Abstract
The pore-forming mechanism of the cholesterol-dependent cytolysins (CDCs) exhibits an absolute requirement for membrane cholesterol. The structural elements of the CDCs that mediate this interaction are not well understood. Three short hydrophobic loops (L1–L3) and a highly conserved undecapeptide sequence at the tip of domain 4 of the CDC structure are known to anchor the CDC to the membrane. It has been thought that the undecapeptide directly mediates the interaction of the CDCs with a cholesterol-rich cell surface. Herein we show that the L1–L3 loops, not the undecapeptide, are responsible for mediating the specific interaction of the CDCs with cholesterol-rich membranes. The membrane insertion of the undecapeptide was uncoupled from membrane binding by the covalent modification of the undecapeptide cysteine thiol. Modification of the cysteine prevented prepore to pore conversion, but did not affect membrane binding, thus demonstrating that undecapeptide membrane insertion follows that of the L1–L3 loops. These studies provide an example of a structural motif that specifically mediates the interaction of a bacterial toxin with a cholesterol-rich membrane.
Keywords: intermedilysin, perfringolysin, toxin, streptolysin, pneumolysin
The cholesterol-dependent cytolysins (CDCs), a large family of related pore-forming toxins, are produced by >20 different species of Gram-positive bacteria (1). The bacteria release these toxins as stable water-soluble monomers that bind to cholesterol-rich membranes. Membrane binding by the monomers initiates a specific sequence of structural changes that promotes their oligomerization on the cell surface and pore formation (2). Two hallmarks of the CDC mechanism are (i) the absolute dependence of its pore-forming mechanism on the presence of membrane cholesterol, and (ii) thiol activation (3, 4). Past studies with perfringolysin O (PFO) and other CDCs showed that they bound directly to cholesterol-rich membranes (5–10) apparently by the cholesterol-rich rafts (11–13). Hence, cholesterol is thought to function as the receptor for the CDCs.
Early studies showed that a highly conserved undecapeptide sequence (ECTGLAWEWWR), also known as the tryptophan-rich region, in domain 4 of the CDC structure (Fig. 1) was important to the CDC mechanism. For the PFO-like CDCs, the thiol-activated or oxidation-sensitive feature results from the reversible oxidation (or modification) of the cysteine thiol group in the undecapeptide (Fig. 1), which is typically the only cysteine present in the primary structure of the secreted CDC. Oxidation of this cysteine inhibits cytolytic activity and may affect CDC binding to membranes (14), although others have suggested that an event subsequent to membrane binding is sensitive to its oxidation (15, 16). Mutations in other undecapeptide residues also affect CDC binding to cholesterol-rich membranes (17, 18). These studies support the concept that the undecapeptide plays a central role in the CDC cytolytic mechanism, but the nature of its contribution remains unclear.
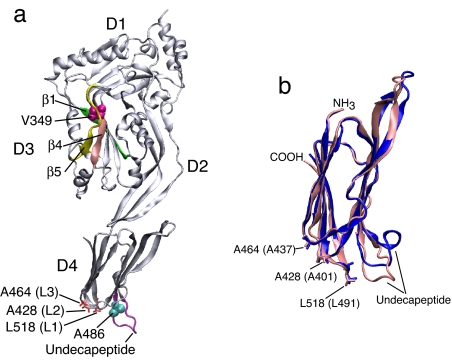
Fig. 1.
The crystal structure of ILY and the domain-4 crystal structures of ILY and PFO. (a) A ribbon representation of the crystal structure of ILY (34) denoting the positions of various structures and residues referred to in this work. (b) An overlay of a ribbon representation of the D4 structures of ILY (pink) and PFO (blue) based on the crystal structures of both proteins (34, 44). Shown are the locations of the undecapeptide and the L1–L3 loop residues of ILY and PFO (the PFO loop residues are in parentheses). The images were generated by using Visual Molecular Dynamics (45).
The generalization that all CDCs use cholesterol as a receptor was complicated by the discovery of a second class of CDCs that binds to a glycoprotein receptor, rather than directly to cholesterol-rich membranes. Streptococcus intermedius intermedilysin (ILY) specifically binds to the surface of human cells (19) by human CD59 (hCD59) (20), a late-stage, species-specific complement inhibitor (21, 22). ILY only binds to hCD59-containing cells, but its cytolytic mechanism remains dependent on the presence of membrane cholesterol (23). ILY can bind to and oligomerize on hCD59-containing cells that have been depleted of cholesterol, but cannot form the membrane pore. Therefore, cholesterol appears to contribute to the ILY cytolytic mechanism in a way unlike it does for most CDCs.
To identify the structural basis for the cholesterol dependence of the pore-forming mechanisms of ILY, a CDC that binds to a nonsterol protein receptor, and PFO, a CDC that binds directly to cholesterol-rich membrane, we performed a detailed study of the interactions between these toxins and cholesterol-rich membranes. These studies revealed a common structural motif in both toxins that specifically mediates their interaction with cholesterol-rich membranes and suggests a unifying explanation for the cholesterol dependence of the PFO and ILY pore-forming mechanisms.
Results
Cholesterol Is Not Required for the Membrane Insertion of ILY Undecapeptide Residue Ala-486.
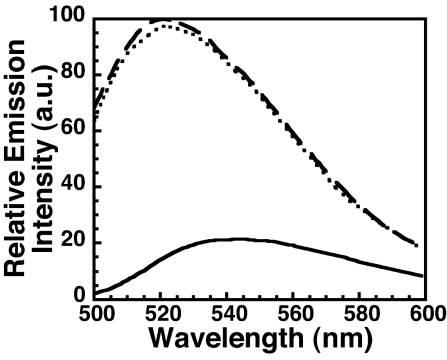
After receptor binding, the first structures of ILY to interact with the membrane are Ala-486 in the D4 undecapeptide and the L1–L3 loops (Fig. 1) (24). Therefore, we determined whether the insertion of Ala-486 required membrane cholesterol. ILYA486C (24) was labeled with the water-sensitive NBD dye, and its fluorescence intensity was measured in the absence and presence of native or cholesterol-depleted membranes. As shown in Fig. 2, the increase in NBD emission intensity in the presence of cholesterol-depleted human RBC (hRBC) ghost membranes reveals that its insertion is not cholesterol-dependent.
Fig. 2.
The ILY undecapeptide residue A486 inserts into cholesterol-depleted membranes. ILY residue Ala-486 was mutated to a cysteine (ILYA486C) and derivatized with NBD. The fluorescence emission of the NBD was determined for ILYA486C-NBD incubated alone (solid line), with hRBCs (dashed line), or with hRBCs depleted of cholesterol (dotted line).
Cholesterol Is Required for the Insertion of ILY Loops L1–L3.
We next examined the membrane insertion of the L1–L3 loops. Insertion of these loops occurs in concert and is required to anchor the CDC monomers on the membrane surface (24, 25). We previously showed that prepore to pore conversion of ILY can be blocked by either depletion of membrane cholesterol or blocking the membrane insertion of the L1–L3 loops (23, 24). These studies suggested that the depletion of membrane cholesterol would block the insertion of the L1–L3 loops and prevent prepore to pore conversion.
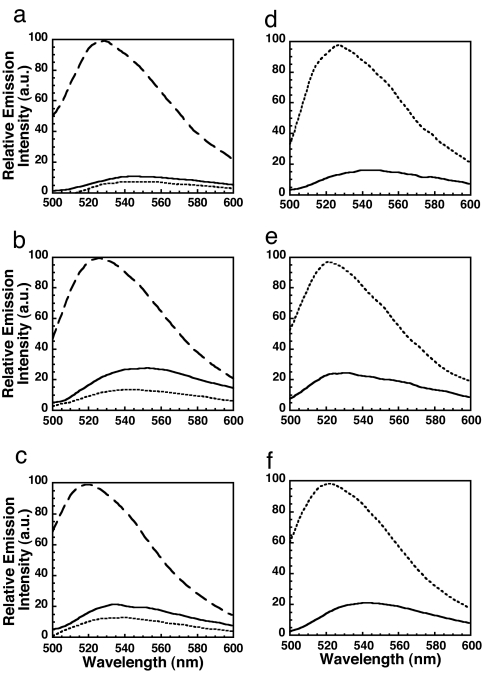
To test this hypothesis, a cysteine was substituted for a residue in each loop (ILYA428C, ILYA464C, and ILYL518C), modified with NBD, and used independently to detect the membrane insertion of each loop into the membrane (24). Whereas the emission intensity of each NBD-labeled loop increased significantly upon binding to native hRBC ghosts, little or no increase in NBD intensity occurred upon hCD59-dependent binding to cholesterol-depleted membranes (Fig. 3 a–c). Thus, depletion of ≈90% of membrane cholesterol prevents the membrane insertion of all three loops. This effect is reversible because restoring cholesterol to the cholesterol-depleted membranes also restored the ability of the loops to insert into the membrane (Fig. 3 d–f). Hence, membrane cholesterol is required for the insertion of the ILY loops L1–L3.
Fig. 3.
L1, L2, and L3 of ILY do not insert into cholesterol-depleted membranes. Each D4 loop was substituted with a cysteine, modified with NBD, and the fluorescence emission was independently determined for each in the absence or presence of native or cholesterol-depleted hRBC ghost membranes. (a–c) ILYA428C-NBD (a), ILYA464C-NBD (b), or ILYL518C-NBD (c) was incubated alone (solid line), with hRBCs (dashed line), or with hRBCs depleted of cholesterol (dotted line). (d–f) Membrane cholesterol was then restored, and insertion was determined for ILYA428C-NBD (d), ILYA464C-NBD (e), or ILYL518C-NBD (f) alone (solid line) or after incubation with cholesterol-replete membranes (dotted line).
Aspartate and Glycine Substitution of Residues in Loops L1–L3 of PFO Prevent its Binding to Cholesterol-Rich Liposomes.
Unlike ILY, PFO binds directly to cholesterol-rich membranes (17, 26, 27) without the aid of a separate receptor. After discovering that the membrane insertion of ILY loops L1–L3 was sensitive to cholesterol, we determined whether these same loops mediated the cholesterol-dependent binding of PFO to liposomes. Thus, the PFO loop residues Ala-401, Ala-437, and Leu-491 (Fig. 1) were individually substituted with aspartate to prevent their insertion into the membrane. We previously showed that substitution of aspartate for the analogous residues in ILY prevented the insertion of its L1–L3 loops and that their insertion was coupled (24). Therefore, we expected that aspartate substitution for any one of the analogous residues in PFO would prevent PFO binding to cholesterol-rich liposomes. The aspartate substitutions, as well as the glycine substitutions described later, did not affect the overall structure of PFO because they exhibited the same trypsin sensitivity as native PFO (data not shown). This finding was not unexpected because these residues project out from the bottom of domain 4 and do not interact with other residues.
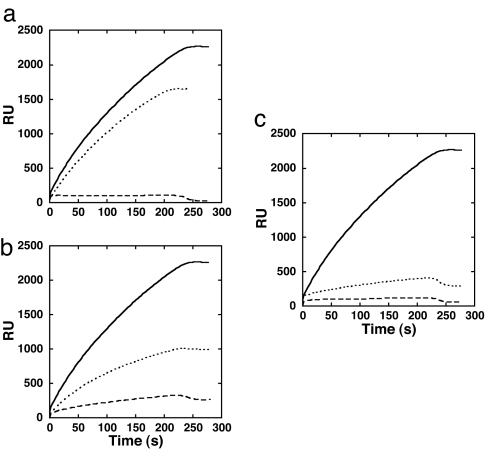
Individual substitution of PFO Ala-401 (L2), Ala-437 (L3), and Leu-491 (L1) with aspartate resulted in a loss of >99% of the hemolytic activity for each mutant (data not shown). Binding of the aspartate-substituted PFO mutants to cholesterol-PC liposomes was then measured by surface plasmon resonance (SPR). Each of these mutations significantly reduced PFO binding to cholesterol-PC liposomes (Fig. 4). Substitution of aspartate for Ala-401 (L2) or Leu-491 (L1) completely blocked the binding of PFO to liposomal membranes. The binding of aspartate-substituted Ala-437 in L3 was <7% of the wild-type binding. These results show that the D4 L1–L3 loops are each required for binding PFO-like CDCs to cholesterol-rich membranes.
Fig. 4.
The L1–L3 loops mediate PFO binding to cholesterol-rich liposomes. Shown is SPR-binding analysis of aspartate- and glycine-substituted PFO loop mutants for residues Ala-401 (loop L2), Ala-437 (loop L3), and Leu-491 (loop L1). (a) SPR-detected binding of native PFO (solid line), PFOA401D (dashed line), and PFOA401G (dotted line). (b) SPR-detected binding of native PFO (solid line), PFOA437D (dashed line), and PFOA437G (dotted line). (c) SPR-detected binding of native PFO (solid line), PFOL491D (dashed line), and PFOL491G (dotted line). RU, resonance units.
Presumably, the structure of the amino acid side chain of these residues is important for recognition of cholesterol-rich membranes. Glycine was substituted for each of these PFO loop residues to evaluate the relative contribution of each side chain to the cholesterol interaction. The substitution of glycine for Ala-401, Ala-437, and Leu-491 of PFO reduced hemolytic activity to 40%, 4%, and 0.4%, respectively, of native PFO. These results are similar to what we found for the analogous glycine substitutions in ILY (data not shown). These substitutions in PFO also decreased its binding to liposomes in a similar fashion (PFOA401G>PFOA437G>PFOL491G) (Fig. 4). Because the loss of the Leu-491 side chain caused the most dramatic reduction in PFO activity and binding, L1 may be most critical in stabilizing PFO interaction with the cholesterol-rich membrane.
Modification of Cys-459 of PFO Does Not Prevent Membrane Binding of PFO.
It has been suggested that oxidation or mutation of the conserved undecapeptide cysteine affects PFO binding to cholesterol-rich membranes, although this theory is controversial (14–16). To resolve this issue, we first compared the binding of native PFO and PFO modified at the Cys-459 sulfhydryl group to cholesterol-rich liposomes by using FRET between donor-labeled toxin and acceptor-labeled liposomes. We then confirmed these results by using SPR.
Modification of the PFO undecapeptide Cys-459 thiol with the sulfhydryl-specific reagent N-ethylmaleimide (NEM) (PFONEM), or with the maleimide derivative of Alexa-488 (PFOAlexa), reduced the hemolytic activity ≥99% (data not shown), similar to other reports in which the sulfhydryl was chemically modified (14, 28). FRET analysis showed that significant acceptor-dependent quenching occurred when PFOAlexa was mixed with rhodamine-labeled liposomes (Fig. 5a), thereby demonstrating that the modified PFO associated with the cholesterol-containing membranes. SPR analysis showed that the rate and extent of PFONEM binding to liposomes were increased relative to native PFO (similar results were obtained with PFOAlexa) (Fig. 5b) (data not shown). Thus, chemical modification of Cys-459 did not block binding of PFO to cholesterol-rich liposomes.
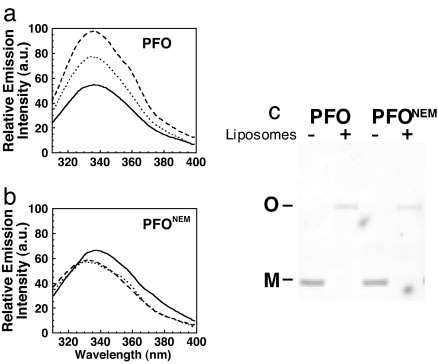
Fig. 5.
Chemical modification of the undecapeptide cysteine of PFO does not prevent binding to cholesterol-rich liposomes. (a) FRET between PFOC459-Alexa and unlabeled liposomes (solid line) or rhodamine-PE-labeled liposomes (dashed line). (b) The SPR-detected binding of native PFO (solid line) and native PFO modified at the native undecapeptide cysteine (Cys-459) with NEM (PFONEM) (dashed line). RU, resonance units.
Modification of PFO Cys-459 Blocks Membrane Insertion of the Undecapeptide Tryptophan Residues and Prevents the Prepore to Pore Transition.
If modification of Cys-459 did not affect PFO membrane binding, how did this modification effectively block pore-forming activity? In addition to the L1–L3 loops, the D4 undecapeptide tryptophan residues also insert into the membrane (27, 29, 30), and their insertion is conformationally coupled to the insertion of the D3 transmembrane β-hairpins (TMHs) (29). The membrane insertion of these tryptophan residues can be monitored by an increase in their intrinsic fluorescence as they move into the membrane (27, 29). Their insertion was measured in PFONEM and native PFO (Fig. 6 a and b). Native PFO exhibited the characteristic increase in fluorescence emission intensity as the undecapeptide tryptophans moved into the membrane. The tryptophan emission was significantly quenched by the incorporation of a nitroxide-labeled lipid into the bilayer, thus demonstrating their presence in the membrane. In contrast, the characteristic increase in the fluorescence emission of the tryptophans was not observed for PFONEM, and the fluorescence emission was not quenched by the nitroxide-labeled phospholipid. Although PFONEM did not form a pore, it bound the membrane and oligomerized into the typical SDS-resistant oligomeric complex seen for native PFO (Fig. 6c). Hence, covalent modification of the undecapeptide cysteine sulfhydryl does not impair the interaction of PFO with cholesterol-rich liposomes, but instead blocks the insertion of the undecapeptide tryptophan residues and prevents prepore to pore conversion.
Fig. 6.
Modification of the PFO undecapeptide cysteine thiol blocks membrane insertion of the undecapeptide tryptophans and conversion of the prepore to pore. (a) The intrinsic emission intensity of the tryptophans in native PFO increases as it moves from solution (solid line) to its membrane-bound state (dashed line). The emission of the tryptophans was quenched by the inclusion of the membrane-restricted collisional quencher 7-DOXYL (dotted line). (b) The experiments shown in a were repeated with native PFO that was modified at Cys-459 with NEM. (c) SDS/AGE analysis of oligomer formation by native PFO and NEM-modified PFO.
Discussion
The studies herein reveal the molecular basis for two characteristic properties of the CDCs (Fig. 7): (i) the absolute dependence of their pore-forming mechanism on cholesterol, and (ii) the inactivation of most CDCs by modification or oxidation of the undecapeptide cysteine. ILY was key to identifying the structural motifs that mediate the cholesterol-dependent interaction of the CDCs with membranes. The ability of ILY to bind first to its protein receptor hCD59 before interacting with lipid allowed us to identify the ILY structural motifs whose membrane insertion was sensitive to the presence of membrane cholesterol. This approach could not be used with PFO because it does not bind to membranes lacking sufficient cholesterol. However, by using receptor-bound ILY, we were able to determine that membrane insertion of the L1–L3 loops was prevented by removing cholesterol from hRBC membranes, and that, in turn, inhibited prepore to pore transition. Because ILY is trapped in the prepore complex by either mutating these loops or depleting membrane cholesterol (23, 24), the cholesterol-dependent insertion of L1–L3 is required for the ILY oligomer to proceed beyond the prepore stage.
Fig. 7.
Schematic summary of the effects on pore formation of cholesterol and oxidation of the undecapeptide cysteine thiol. ILY and PFO D4 domains are depicted with membranes (gray). (Upper) After ILY binds to hCD59, the L1–L3 loops insert into the membrane in a cholesterol-dependent manner. This insertion is followed by the cholesterol-independent insertion of the undecapeptide residue Ala-486 of the undecapeptide with the subsequent formation of the pore. It is not known whether the tryptophan residues of the ILY undecapeptide insert into the membrane (they are not shown as inserted in the model). In the absence of cholesterol, Ala-486 of ILY inserts into the membrane after receptor binding, but loops L1–L3 do not, thus trapping ILY in the prepore complex. (Lower) Loops L1–L3, the undecapeptide tryptophan residues, and Cys-459 of PFO insert into the membrane in cholesterol-rich membranes. Preventing the insertion of any single L1–L3 loop prevents PFO binding to cholesterol-rich membranes, similar to the effect seen with membranes that lack cholesterol. Oxidation or modification of the Cys-459 thiol blocks membrane insertion of the undecapeptide tryptophan residues and traps PFO in the prepore complex.
Because of the highly conserved primary structure of the undecapeptide, and the fact that the undecapeptide tryptophan residues insert into the membrane surface (27, 29), the undecapeptide was widely assumed to mediate the interaction of the PFO and PFO-like CDCs with cholesterol-rich membranes. However, the studies herein show that binding and oligomerization of PFO, a CDC that binds directly to cholesterol-rich membranes, are independent of undecapeptide tryptophan membrane insertion. Covalent modification of the Cys-459 thiol group uncoupled the insertion of the undecapeptide tryptophans from the insertion of the L1–L3 loops without decreasing the extent of binding or oligomerization of PFO. In contrast, disrupting the insertion of any one of the L1–L3 loops blocked its binding to cholesterol-rich membranes. These data showed that only the L1–L3 loops were necessary for PFO binding to the cholesterol-rich membrane. It is unlikely that other regions of D4 participate in this interaction because we previously showed that it is oriented perpendicular to the membrane surface and, with the exception of the L1–L3 loops and undecapeptide, D4 is surrounded by water (25). Also, domains 1–3 of PFO are positioned ≥40Å above the membrane surface and only come into close proximity to the membrane surface upon conversion of the prepore oligomer to the pore complex (31, 32) and so do not participate in membrane binding. Thus, the L1–L3 loops mediate the initial direct interaction of PFO-like CDCs with a cholesterol-rich membrane, whereas in ILY L1–L3 only insert in a cholesterol-dependent fashion after ILY binds to hCD59.
These studies also provide a molecular basis for the hallmark trait of thiol-activated CDCs. Small thiols in crude preparations of the CDCs apparently form disulfides with the undecapeptide cysteine over time and cause a loss of activity that can be reversed by reducing agents (1). As described earlier, chemical modification of the thiol group of the PFO undecapeptide cysteine blocked the membrane insertion of the D4 undecapeptide tryptophan residues. Membrane insertion of these residues precedes but is conformationally coupled to membrane insertion of the D3 TMHs that form the transmembrane β-barrel pore (29). Hence, modification of Cys-459 appears to block the conformational changes that position the D4 tryptophans in the membrane and disrupts the conformationally coupled pathway that triggers insertion of the D3 TMHs.
It is interesting that the L1–L3 loops do not facilitate the direct binding of ILY to cholesterol-rich membranes as they do for the PFO and PFO-like CDCs. The ILY L1–L3 loops only insert into the membrane in a cholesterol-dependent fashion after ILY binds to its hCD59 receptor. This difference may result from differences in the structure of the undecapeptide in ILY and PFO. Nagamune et al. (33) showed that replacement of the ILY undecapeptide (GATGLAWEPWR) with that of the consensus undecapeptide (ECTGLAWEWWR) allowed ILY to bind to nonhuman cells. This result suggested that the structure of the undecapeptide was an important factor in the toxin–membrane interaction. Yet as we have shown herein, the undecapeptide is not directly involved in mediating binding of PFO-like CDCs with cholesterol-rich membranes. How then does the undecapeptide structure influence the interaction of the CDCs with cholesterol-rich membranes?
The recent studies of Soltani et al. (24) may provide some insight into the role of the undecapeptide in membrane binding. They showed that mutation of the ILY undecapeptide Trp-491 to alanine prevented the insertion of the L1–L3 loops, thereby demonstrating that the membrane insertion of the L1–L3 loops depends on the conformation of the undecapeptide. Indeed, the D4 structures of the soluble monomers of PFO and ILY are nearly identical (rms deviation of <0.6 Å) (34), including the location and orientation of the L1–L3 loop residues (Fig. 1b). The only major deviations in their 3D structures are the undecapeptide loop at the bottom of D4 and a β-tongue structure at the top of D4. As shown here, the ILY L1–L3 loops insert into cholesterol-rich membranes after receptor binding. Presumably, receptor binding by ILY alters the conformation of the undecapeptide region so insertion of the L1–L3 loops can occur. Hence, although the undecapeptide does not directly mediate the interaction of ILY and other CDCs with cholesterol-rich membranes, its structure may be a critical modulator of this interaction.
Do the L1–L3 loop residues constitute a cholesterol-binding site for the CDCs? Our data show that L1–L3 specifically interact with cholesterol-rich membranes to trigger pore formation by ILY and mediate PFO binding. Therefore, it is clear that the L1–L3 loops either contact one or more cholesterol molecules directly and/or associate with a membrane surface that has specific cholesterol-dependent characteristics. Either way, the next goal is to identify the specific residues in L1–L3 that are involved in cholesterol recognition.
In summary, this study revealed roles of certain CDC structural elements in two long-standing hallmarks of the CDC pore-forming mechanism: (i) the sensitivity of pore formation to membrane cholesterol, and (ii) the oxidation or modification of the undecapeptide cysteine thiol. The results reported here also may guide future studies of protein–membrane interactions for other toxins (35–40) and viruses (41) that exhibit a cholesterol-dependent mechanism.
Methods
Bacterial Strains, Plasmids, and Chemicals.
The genes for ILY and PFO were cloned into pTrcHisA (Invitrogen) as described previously (23, 42). All mutations were made in the native ILY (naturally cysteine-less), native PFO, or cysteine-less PFO (PFOC459A) background. Native PFO has not been mutated and contains a cysteine at residue 459. Both PFO and PFOC459A exhibit similar cytolytic activities (42). All chemicals and enzymes were obtained from Sigma–Aldrich, VWR, and Research Organics. All fluorescent probes were obtained from Molecular Probes (Invitrogen).
Generation, Purification, and Trypsin Sensitivity of Toxins and Their Derivatives.
By using PCR QuikChange mutagenesis (Stratagene), various amino acid substitutions were made in native ILY, PFO, or the functional cysteine-less derivative of PFO, PFOC459A (42). The Oklahoma Medical Research Foundation Core DNA Sequencing Facility performed DNA sequence analysis. The expression and purification of recombinant ILY and its derivatives from Escherichia coli were carried out as described (24, 42). Purified protein was dialyzed into buffer [300 mM NaCl, 10 mM Mes, 1 mM EDTA (pH 6.5)] overnight at 4°C and stored in 5 mM DTT and 10% (vol/vol) sterile glycerol at −80°C.
Each ILY and PFO mutant was analyzed for perturbations in its structure by comparing the trypsin sensitivity of each mutant to that of native toxin as previously described (24).
Chemical Modification of ILY and PFO and Their Derivatives with Sulfhydryl-Specific Probes.
The cysteine derivatives were modified with the environmentally sensitive probe iodoacetamido-N,N′-dimethyl-N-(7-nitrobenz-2-oxa-1,3-diazolyl)ethylene-diamine (NBD) by the sulfhydryl group. The reaction was carried out as previously described (23). Modified protein was stored in 10% (vol/vol) sterile glycerol, quick frozen in liquid nitrogen, and stored at −80°C. Proteins were labeled at an efficiency of ≥75%.
PFO was labeled at the native cysteine (residue 459) with NEM (Sigma–Aldrich). In a typical reaction, 1 mg of PFO was incubated with a 20-fold molar excess of NEM for 30 min. The reaction was then passed over a Sephadex G-50 equilibrated in Hepes-buffered saline (HBS) [100 mM NaCl, 50 mM Hepes (pH 8.0)] to separate labeled toxin from the excess NEM. The modified PFO was stored on ice and used within 3 days.
Liposome Preparation.
Liposomes containing 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and cholesterol (Avanti Polar Lipids) at a molar ratio of 45:55 were prepared as described (42). Rhodamine phosphotidylethanolamine (PE)-labeled liposomes were prepared in a similar fashion, except that 10 mol% of the total lipid was substituted with rhodamine PE. Liposomes that contained a membrane-restricted nitroxide collisional quencher were prepared by substituting 10 mol% of the total lipid with 1-palmitoyl-2-stearoyl-(7-DOXYL)-sn-glycero-3-phosphocholine (i.e., 7-DOXYL).
Fluorescence Measurements.
All fluorescence intensity measurements were performed by using an SLM-8100 photon-counting spectrofluorimeter as previously described (42). For NBD measurements, an excitation wavelength of 470 nm was used (bandpass of 4 nm). Emission scans from 500–600 nm were recorded for each sample at a resolution of 1 nm and an integration time of 1 s. Samples containing 0.18 nmol of ILY were incubated with hRBC ghost membranes (equivalent to ≈300 μg of membrane protein) in PBS [10 mM Na2HPO4, 2 mM KH2PO4 (pH 7.5), 137 mM NaCl, 3 mM KCl] at 37°C for 10 min before making spectral measurements.
FRET between 0.18 nmol PFO labeled at its native cysteine (Cys-459) with Alexa 488 and rhodamine-PE-labeled liposomes was carried out under conditions described above for the NBD experiments, except that the ghost membranes were replaced by either liposomes or PE-rhodamine-containing liposomes (82 μM lipid in a 2-ml sample volume).
Intrinsic tryptophan emission intensity of PFO was measured as described earlier for the NBD studies, except that the emission wavelength was recorded between 300 and 400 nm and the excitation wavelength was set to 270 nm (4-nm bandpass). The emission intensity was recorded in the absence and presence of POPC:cholesterol liposomes, and the same liposomes that had 10 mol% of the total lipid replaced with 7-DOXYL.
hRBC Ghost Membrane Preparation.
hRBC ghost membranes were prepared as previously described (23, 42). Membrane protein content was quantified by using the Bradford method (Bio-Rad protein assay) (23, 42).
Cholesterol Depletion and Repletion of hRBC Ghost Membranes.
Cholesterol extraction was performed with methyl-β-cyclodextrin (MβCD) as previously described (23). Briefly, hRBC ghost membranes were incubated with a final concentration of 20–40 mM MβCD (made fresh for each use) at 37°C for 2 h. The membranes were washed three times by repeated centrifugation (15,700 × g for 20 min at 4°C) and resuspended in PBS to remove excess MβCD. Ghost membranes were finally suspended in PBS. Cholesterol content was measured by using Cholesterol/Cholesteryl Ester Quantitation Kit (Calbiochem). Typically, the cholesterol content of the membranes was decreased >90% by this method.
Cholesterol repletion was performed by using cholesterol-loaded MβCD as previously described (23). Briefly, MβCD was added to buffer A [140 mM NaCl, 5 mM KCl, 5 mM KH2PO4 (pH 6.5), 1 mM MgSO4, 10 mM Hepes, 5 mM glucose] to a final concentration of 5 mM. MβCD in buffer A was heated to 80°C in a glass container, and a cholesterol suspension [100 mM cholesterol in 1: 2 (vol/vol) chloroform:methanol] was added to a final concentration of 4 mM. The solution was clarified by sonication (4 × 20 s). The solution was filtered by using a 0.22-μm filter. MβCD loaded with cholesterol (4 mM final cholesterol concentration) was added to pelleted cholesterol-depleted ghost membranes (equivalent to ≈4 mg/ml membrane protein) and incubated for 2 h at 37°C. The membranes were washed by repeated centrifugation as before and resuspended in PBS at a concentration of 3–5 mg membrane protein per ml.
Immobilization of Liposomes on L1 SPR Sensor Chip.
SPR was measured with a BIAcore 3000 system by using a L1 sensor chip (BIAcore). The L1 sensor chip contains a dextran matrix to which hydrophobic residues are covalently bound and used for immobilization of liposomes. To prepare the L1 chip for liposomes, 10 μl of 20 mM CHAPS were injected at a flow rate of 10 μl/min. Liposomes (0.5 mM final lipid concentration) were then injected at the same flow rate for 10 min. After injection of liposomes, 50 mM NaOH was injected for 3 min to remove the multiple layers of lipids, followed by an injection of 0.1 mg/ml BSA to coat the nonspecific binding sites. All injections were performed at 25°C. The L1 chip was regenerated and stripped of liposomes by repeated injections of 20 mM CHAPS and 50 mM NaOH until original RU reading was reached. No loss of sensor chip-binding capacity resulted from regeneration.
SPR Analysis.
Analysis of liposome–PFO (and PFO derivatives') interactions were performed in HBS at 25°C. PFO and its derivatives were injected over the liposome-coated chip at a concentration of 50 ng/μl and a flow rate of 30 μl/min for 4 min.
SDS/Agarose Gel Electrophoresis (AGE).
SDS/AGE was carried out as in ref. 43.
ACKNOWLEDGMENTS.
This work was supported by National Institutes of Health Grant AI037657 and Robert A. Welsh Foundation Grant BE-0017.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Alouf JE, Billington SJ, Jost BH. In: Bacterial Toxins: A Comprehensive Sourcebook. Alouf JE, Popoff MR, editors. London: Academic; 2005. pp. 643–658. [Google Scholar]
- 2.Tweten RK. Infect Immun. 2005;73:6199–6209. doi: 10.1128/IAI.73.10.6199-6209.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernheimer AW. J Bacteriol. 1966;91:1677–1680. doi: 10.1128/jb.91.5.1677-1680.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernheimer AW, Davidson M. Science. 1965;148:1229–1231. doi: 10.1126/science.148.3674.1229. [DOI] [PubMed] [Google Scholar]
- 5.Shany S, Bernheimer AW, Grushoff PS, Kim KS. Mol Cell Biochem. 1974;3:179–186. doi: 10.1007/BF01686643. [DOI] [PubMed] [Google Scholar]
- 6.Badin J, Denne MA. Cell Mol Biol. 1978;23:133–143. [PubMed] [Google Scholar]
- 7.Delattre J, Lebsir R, Panouse PJ, Badin J. Cell Mol Biol. 1979;24:157–166. [PubMed] [Google Scholar]
- 8.Johnson MK, Geoffroy C, Alouf JE. Infect Immun. 1980;27:97–101. doi: 10.1128/iai.27.1.97-101.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rottem S, Cole RM, Habig WH, Barile MF, Hardegree MC. J Bacteriol. 1982;152:888–892. doi: 10.1128/jb.152.2.888-892.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohno-Iwashita Y, Iwamoto M, Mitsui K, Ando S, Nagai Y. Eur J Biochem. 1988;176:95–101. doi: 10.1111/j.1432-1033.1988.tb14255.x. [DOI] [PubMed] [Google Scholar]
- 11.Ohsaki Y, Sugimoto Y, Suzuki M, Kaidoh T, Shimada Y, Ohno-Iwashita Y, Davies JP, Ioannou YA, Ohno K, Ninomiya H. Histochem Cell Biol. 2004;121:263–272. doi: 10.1007/s00418-004-0643-7. [DOI] [PubMed] [Google Scholar]
- 12.Shimada Y, Maruya M, Iwashita S, Ohno-Iwashita Y. Eur J Biochem. 2002;269:6195–6203. doi: 10.1046/j.1432-1033.2002.03338.x. [DOI] [PubMed] [Google Scholar]
- 13.Waheed AA, Shimada Y, Heijnen HF, Nakamura M, Inomata M, Hayashi M, Iwashita S, Slot JW, Ohno-Iwashita Y. Proc Natl Acad Sci USA. 2001;98:4926–4931. doi: 10.1073/pnas.091090798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwamoto M, Ohno-Iwashita Y, Ando S. Eur J Biochem. 1987;167:425–430. doi: 10.1111/j.1432-1033.1987.tb13355.x. [DOI] [PubMed] [Google Scholar]
- 15.Saunders FK, Mitchell TJ, Walker JA, Andrew PW, Boulnois GJ. Infect Immun. 1989;57:2547–2552. doi: 10.1128/iai.57.8.2547-2552.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vazquez-Boland JA, Dominguez L, Rodriguez-Ferri EF, Fernandez-Garayzabal JF, Suarez G. FEMS Microbiol Lett. 1989;53:95–99. doi: 10.1016/0378-1097(89)90372-8. [DOI] [PubMed] [Google Scholar]
- 17.Sekino-Suzuki N, Nakamura M, Mitsui KI, Ohno-Iwashita Y. Eur J Biochem. 1996;241:941–947. doi: 10.1111/j.1432-1033.1996.00941.x. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs T, Cima-Cabal MD, Darji A, Mendez FJ, Vazquez F, Jacobs AA, Shimada Y, Ohno-Iwashita Y, Weiss S, de los Toyos JR. FEBS Lett. 1999;459:463–466. doi: 10.1016/s0014-5793(99)01297-1. [DOI] [PubMed] [Google Scholar]
- 19.Nagamune H, Ohnishi C, Katsuura A, Fushitani K, Whiley RA, Tsuji A, Matsuda Y. Infect Immun. 1996;64:3093–3100. doi: 10.1128/iai.64.8.3093-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giddings KS, Zhao J, Sims PJ, Tweten RK. Nat Struct Mol Biol. 2004;12:1173–1178. doi: 10.1038/nsmb862. [DOI] [PubMed] [Google Scholar]
- 21.Rollins SA, Zhao J, Ninomiya H, Sims PJ. J Immunol. 1991;146:2345–2351. [PubMed] [Google Scholar]
- 22.Rollins SA, Sims PJ. J Immunol. 1990;144:3478–3483. [PubMed] [Google Scholar]
- 23.Giddings KS, Johnson AE, Tweten RK. Proc Natl Acad Sci USA. 2003;100:11315–11320. doi: 10.1073/pnas.2033520100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soltani CE, Hotze EM, Johnson AE, Tweten RK. J Biol Chem. 2007;282:15709–15716. doi: 10.1074/jbc.M701173200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramachandran R, Heuck AP, Tweten RK, Johnson AE. Nat Struct Biol. 2002;9:823–827. doi: 10.1038/nsb855. [DOI] [PubMed] [Google Scholar]
- 26.Iwamoto M, Ohno-Iwashita Y, Ando S. Eur J Biochem. 1990;194:25–31. doi: 10.1111/j.1432-1033.1990.tb19422.x. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura M, Sekino N, Iwamoto M, Ohno-Iwashita Y. Biochemistry. 1995;34:6513–6520. doi: 10.1021/bi00019a032. [DOI] [PubMed] [Google Scholar]
- 28.Harris JR, Adrian M, Bhakdi S, Palmer M. J Struct Biol. 1998;121:343–355. doi: 10.1006/jsbi.1998.3989. [DOI] [PubMed] [Google Scholar]
- 29.Heuck AP, Hotze E, Tweten RK, Johnson AE. Molec Cell. 2000;6:1233–1242. doi: 10.1016/s1097-2765(00)00119-2. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura M, Sekino-Suzuki N, Mitsui K, Ohno-Iwashita Y. J Biochem (Tokyo) 1998;123:1145–1155. doi: 10.1093/oxfordjournals.jbchem.a022054. [DOI] [PubMed] [Google Scholar]
- 31.Czajkowsky DM, Hotze EM, Shao Z, Tweten RK. EMBO J. 2004;23:3206–3215. doi: 10.1038/sj.emboj.7600350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramachandran R, Tweten RK, Johnson AE. Proc Natl Acad Sci USA. 2005;102:7139–7144. doi: 10.1073/pnas.0500556102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagamune H, Ohkura K, Sukeno A, Cowan G, Mitchell TJ, Ito W, Ohnishi O, Hattori K, Yamato M, Hirota K, et al. Microbiol Immunol. 2004;48:677–692. doi: 10.1111/j.1348-0421.2004.tb03479.x. [DOI] [PubMed] [Google Scholar]
- 34.Polekhina G, Giddings KS, Tweten RK, Parker MW. Proc Natl Acad Sci USA. 2005;102:600–605. doi: 10.1073/pnas.0403229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishitsuka R, Kobayashi T. Biochemistry. 2007;46:1495–1502. doi: 10.1021/bi061290k. [DOI] [PubMed] [Google Scholar]
- 36.Tomita T, Noguchi K, Mimuro H, Ukaji F, Ito K, Sugawara-Tomita N, Hashimoto Y. J Biol Chem. 2004;279:26975–26982. doi: 10.1074/jbc.M402676200. [DOI] [PubMed] [Google Scholar]
- 37.Barlic A, Gutierrez-Aguirre I, Caaveiro JM, Cruz A, Ruiz-Arguello MB, Perez-Gil J, Gonzalez-Manas JM. J Biol Chem. 2004;279:34209–34216. doi: 10.1074/jbc.M313817200. [DOI] [PubMed] [Google Scholar]
- 38.Chattopadhyay K, Bhattacharyya D, Banerjee KK. Eur J Biochem. 2002;269:4351–4358. doi: 10.1046/j.1432-1033.2002.03137.x. [DOI] [PubMed] [Google Scholar]
- 39.Giesemann T, Jank T, Gerhard R, Maier E, Just I, Benz R, Aktories K. J Biol Chem. 2006;281:10808–10815. doi: 10.1074/jbc.M512720200. [DOI] [PubMed] [Google Scholar]
- 40.Martin C, Requero MA, Masin J, Konopasek I, Goni FM, Sebo P, Ostolaza H. J Bacteriol. 2004;186:3760–3765. doi: 10.1128/JB.186.12.3760-3765.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kielian M, Chatterjee PK, Gibbons DL, Lu YE. Subcell Biochem. 2000;34:409–455. doi: 10.1007/0-306-46824-7_11. [DOI] [PubMed] [Google Scholar]
- 42.Shepard LA, Heuck AP, Hamman BD, Rossjohn J, Parker MW, Ryan KR, Johnson AE, Tweten RK. Biochemistry. 1998;37:14563–14574. doi: 10.1021/bi981452f. [DOI] [PubMed] [Google Scholar]
- 43.Shepard LA, Shatursky O, Johnson AE, Tweten RK. Biochemistry. 2000;39:10284–10293. doi: 10.1021/bi000436r. [DOI] [PubMed] [Google Scholar]
- 44.Rossjohn J, Feil SC, McKinstry WJ, Tweten RK, Parker MW. Cell. 1997;89:685–692. doi: 10.1016/s0092-8674(00)80251-2. [DOI] [PubMed] [Google Scholar]
- 45.Humphrey W, Dalke A, Schulten K. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]