Abstract
The spindle checkpoint ensures the accuracy of chromosome segregation during mitosis. The protein serine/threonine kinase, Mps1, is a critical component of the spindle checkpoint in human cells and regulates the kinetochore localization of key checkpoint proteins. The kinase activity of Mps1 is required for the spindle checkpoint, but how Mps1 is activated during mitosis is unclear. Here, we show that the endogenous Mps1 in mitotic HeLa cells is phosphorylated on T676, a residue in the activation loop. This phosphorylation event on Mps1 is required for its kinase activity in vitro and for spindle checkpoint signaling in vivo. T676 phosphorylation of Mps1 increases during mitosis and can occur through intermolecular/trans autophosphorylation. Induced dimerization of Mps1 is sufficient to activate its kinase activity in cells. We speculate that the kinetochore localization of Mps1 raises its local concentration, leading to its activation during mitosis through more efficient trans autophosphorylation.
Keywords: mitosis, kinase, kinetochore, activation loop
Accurate chromosome segregation during mitosis is critical for maintaining genomic stability (1). A cell-cycle surveillance system called the spindle checkpoint ensures faithful chromosome segregation by delaying anaphase onset until all sister chromatids achieve proper attachment to the mitotic spindle (2–4). A single unattached kinetochore is sufficient to activate the spindle checkpoint. This high sensitivity of the spindle checkpoint suggests that certain steps of checkpoint signaling need to be catalytic. Indeed, multiple spindle checkpoint proteins are protein serine/threonine kinases, including Aurora B, Mps1, Bub1, and BubR1 (2–4). These kinases are recruited to kinetochores, the originating sites of checkpoint signaling, during mitosis. They likely form kinase cascades to transduce and amplify checkpoint signals from the kinetochores in ways analogous to the MAP kinase cascades in growth factor-dependent signal transduction pathways (2–5).
MPS1 was first identified in budding yeast as a gene required for spindle pole body (SPB) duplication (6). It was later shown that Mps1 is crucial for spindle checkpoint signaling in yeast (7). Overexpression of Mps1 in yeast is sufficient to activate the spindle checkpoint and arrests cells in mitosis in the absence of spindle damage (8). Subsequent studies then identified orthologs of Mps1 in various organisms, revealing that the function of Mps1 in the spindle checkpoint is conserved from yeast to man (9–12). In addition, Mps1 has meiotic and developmental functions in multicellular organisms, such as fly and zebra fish (13–17). Mps1 has also recently been reported to play a role in DNA damage response and Smad signaling in human cells (18, 19).
In the spindle checkpoint, Mps1 is required for the kinetochore localization of other checkpoint proteins, including Bub1, BubR1, Mad1, and Mad2 (10, 20). The kinase activity of Mps1 is required for the spindle checkpoint (21, 22). Human Mps1 is phosphorylated and activated during mitosis (11). It is, however, unclear how Mps1 is activated during mitosis. Very recently, bacterially expressed human Mps1 has been shown to undergo autophosphorylation at an activation loop residue, T676 (23). The Mps1 T676A mutant has much lower kinase activity in vitro and, when overexpressed in human cells, fails to cause centrosome overduplication (23).
In this study, we have mapped the phosphorylation sites of the endogenous Mps1 isolated from mitotic human cells by using mass spectrometry. We show that human Mps1 is indeed phosphorylated in vivo at T676. The Mps1 T676A mutant is less active in vitro and fails to restore the mitotic arrest of Mps1 RNAi cells treated with nococazole. Phosphorylation of T676 of Mps1 increases in mitosis, suggesting that it might be an important mechanism for Mps1 activation. Induced dimerization of Mps1 is sufficient to activate Mps1 in cells. Because Mps1 localizes to kinetochores during mitosis, we speculate that high local concentrations of Mps1 at kinetochores might promote its trans autophosphorylation, resulting in its activation.
Results
Identification of in Vivo Mitotic Phosphorylation Sites on Human Mps1.
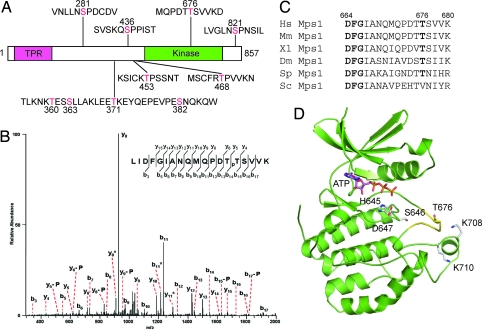
Human Mps1 is hyperphosphorylated, and its kinase activity is elevated during mitosis (11). To understand the activation mechanism of Mps1, we immunoprecipitated (IPed) the endogenous Mps1 from nocodazole-arrested mitotic HeLa cells and analyzed the phosphorylation sites on Mps1 by mass spectrometry. The majority of the expected tryptic peptides of Mps1 were recovered, resulting in 81% coverage of the human Mps1 sequence. A total of 10 phosphorylation sites were identified (Fig. 1 A and B). Five phospho-residues, S281, S436, T453, T468, and S821, were followed by a proline, suggesting that their phosphorylation is mediated by proline-directed kinases, such as cyclin-dependent kinases (CDKs) and MAP kinases (MAPKs). Indeed, S844 in Xenopus Mps1, the residue equivalent to S821 in human Mps1, is phosphorylated by MAPKs (24). This phosphorylation is required for proper kinetochore localization of Mps1 in Xenopus egg extracts (24). In addition, S436, T453, and T468 of an Mps1 fragment are phosphorylated by cyclin E/Cdk2 or cyclin A/Cdk2 in vitro (25).
Fig. 1.
Identification of phosphorylation sites in human Mps1 protein. (A) In vivo phosphorylation sites of Mps1 from mitotic HeLa cells identified by mass spectrometry. (B) Tandem mass spectrum of the phospho-T676-containing peptide. “b” and “y” ion series represent fragment ions containing the N- and C-termini of the peptide, respectively. Ions labeled with “-P” indicate a neutral loss of H3PO4, whereas ions labeled with “*” indicate a neutral loss of NH3. (C) Sequence alignment of the activation loops of Mps1 proteins from Homo sapiens (Hs), Mus musculus (Mm), Xenopus laevis (Xl), Drosophila melanogaster (Dm), Schizosaccharomyces pombe (Sp), and Saccharomyces cerevisiae (Sc). T676 and the DFG motif that binds Mg2+-ATP are shown in bold. (D) A structure model of the kinase domain of human Mps1. The activation loop is shown in yellow. ATP and several residues discussed in the text are shown as sticks.
T676 Phosphorylation Is Required for the Kinase Activity of Mps1.
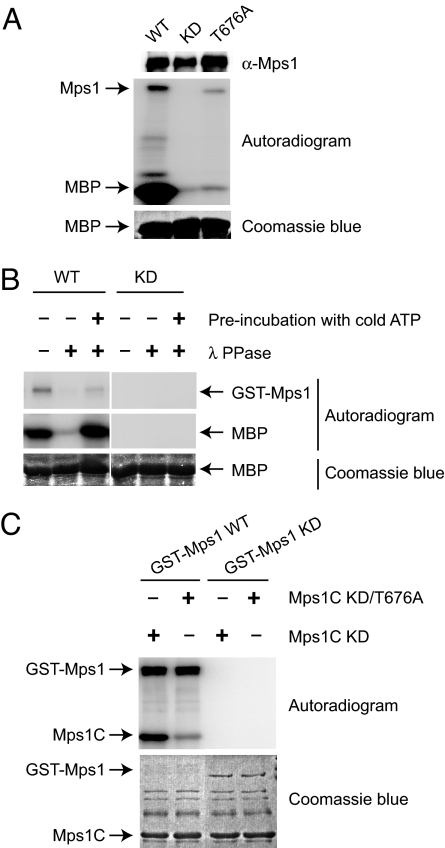
The T676 site is conserved in Mps1 proteins from all organisms examined with the exception of budding yeast [Fig. 1C and supporting information (SI) Fig. 7]. This site is located in the activation loop of Mps1 (Fig. 1D and SI Fig. 7). Phosphorylation of activation loop residues has been shown to be an important activation mechanism for many protein kinases (26). We thus mutated T676 in human Mps1 to alanine and examined the kinase activity of the T676A mutant. For comparison, we also created a kinase-dead (KD) mutant of Mps1 by mutating D664 in the “DFG” motif to alanine. Next, we in vitro translated Myc-Mps1 wild type (WT), KD, and T676A in rabbit reticulocyte lysate, IPed these Myc-Mps1 proteins by using anti-Myc beads, and assayed their kinase activities by using myelin basic protein (MBP) as the substrate. As expected, Myc-Mps1 WT phosphorylated MBP efficiently whereas Mps1 KD was inactive (Fig. 2A). Furthermore, Myc-Mps1 WT, but not KD, also underwent autophosphorylation (Fig. 2A). The kinase activity of Myc-Mps1 T676A toward MBP or itself was much lower than that of Mps1 WT, suggesting that phosphorylation of T676 was required for the kinase activity of Mps1 in vitro.
Fig. 2.
Autophosphorylation of T676 is required for the kinase activity of Mps1. (A) In vitro translated Myc-Mps1 was IPed by using anti-Myc beads and incubated with 5 μg of MBP in the presence of γ-[32P]ATP for 30 min at 30°C. The reaction mixture was separated on SDS/PAGE followed by blotting with anti-Mps1 (Top), autoradiography (Middle), and Coomassie blue staining (Bottom). (B) Approximately 60 ng of the wild-type (WT) or 300 ng of kinase-dead mutant (KD) of GST-Mps1 IPed from Sf9 cell lysate was either untreated or treated with λ phosphatase and assayed for its kinase activity against 5 μg of MBP with or without a preincubation with cold ATP. The reaction mixture was separated on SDS/PAGE and analyzed by autoradiography (Top and Middle) and Coomassie blue staining (Bottom). (C) Approximately 60 ng of the WT or 300 ng of KD of GST-Mps1 was IPed from Sf9 cell lysate by using anti-Mps1 beads and incubated with 2 μg of bacterially expressed C-terminal fragment of Mps1 (Mps1C) in the presence of γ-[32P]ATP for 30 min at 30°C. The reaction mixture was separated on SDS/PAGE and analyzed by autoradiography (Upper) and Coomassie blue staining (Lower).
We next tested whether the T676D or T676E mutations could mimic phosphorylation of T676 in Mps1. Both Mps1 T676D and T676E had much lower kinase activity as compared with Mps1 WT (data not shown), indicating that these mutations could not effectively mimic phosphorylation at T676. It is thus formally possible that T676 is required for the structural integrity of Mps1, and the T676A mutant is inactive for structural reasons instead of the lack of phosphorylation. If so, the T676D and T676E mutations are expected to be more deleterious than the T676A mutation. We note, however, that the kinase activities of Mps1 T676D and T676E were higher than those of Mps1 T676A (data not shown). Therefore, although we cannot rule out a structural role of T676, our results are more consistent with the possibility that phosphorylation of T676 is required for the kinase activity of Mps1.
Phosphorylation of T676 in Mps1 Occurs Through Trans Autophosphorylation.
If phosphorylation of T676 is indeed required for the kinase activity of Mps1, it might occur through autophosphorylation, because Mps1 translated in rabbit reticulocyte lysate is active. To further test whether Mps1 underwent autophosphorylation at T676, we expressed and purified GST-Mps1 WT and KD in Sf9 insect cells. Mass spectrometry analysis of purified GST-Mps1 WT identified T676 and S821 as two major phosphorylation sites (data not shown). Furthermore, dephosphorylation of GST-Mps1 by λ-phosphatase greatly diminished the kinase activity of Mps1 toward MBP (Fig. 2B). A preincubation of phosphatase-treated Mps1 with cold ATP restored its kinase activity (Fig. 2B). Our results suggest that Mps1 undergoes autophosphorylation at T676, which further enhances its kinase activity.
It remains possible that an unknown kinase from Sf9 insect cells is responsible for phosphorylating GST-Mps1 in the above experiments. In fact, phosphorylation at S821 of GST-Mps1 purified from Sf9 cells is very likely mediated by Sf9 CDKs or MAPKs. To further confirm that Mps1 can undergo autophosphorylation at T676, we expressed and purified the kinase-dead mutants of the C-terminal kinase domain of Mps1 (Mps1C KD) in bacteria and used it as substrate in the in vitro kinase assay. GST-Mps1 WT, but not GST-Mps1 KD, efficiently phosphorylated Mps1C KD (Fig. 2C). We next expressed and purified a KD/T676A double mutant of Mps1C from bacteria. GST-Mps1 WT only weakly phosphorylated Mps1C KD/T676A, suggesting that T676 was a major site of phosphorylation (Fig. 2C). These results indicate that Mps1 can undergo autophosphorylation at T676 in trans, i.e., one molecule of Mps1 can phosphorylate another molecule of Mps1 at T676. The efficient autophosphorylation of GST-Mps1 could be due to GST fusion proteins being intrinsically dimeric. During the course of our work, Mattison et al. (23) showed that bacterially expressed Mps1 also underwent autophosphorylation at T676.
Phosphorylation of T676 in Mps1 Is Required for the Spindle Checkpoint.
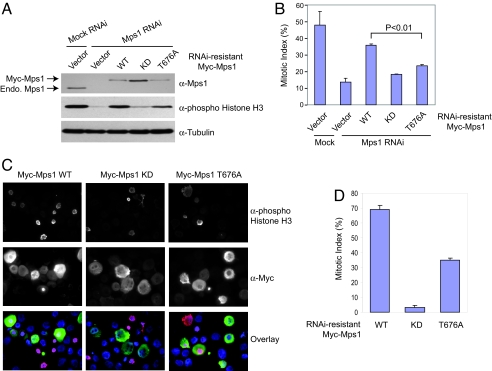
To test whether phosphorylation of T676 in Mps1 is required for the spindle checkpoint, we depleted the endogenous Mps1 from HeLa cells by using RNA interference (RNAi) and compared the ability of ectopically expressed Myc-Mps1 WT, KD, or T676A to restore the spindle checkpoint. HeLa cells have a functional spindle checkpoint and undergo mitotic arrest in the presence of the microtubule-depolymering drug nocodazole. Mps1 RNAi efficiently knocked down the level of Mps1 in HeLa cells (Fig. 3A). Mps1 RNAi cells failed to arrest in mitosis in the presence of nocodazole, based on the levels of the mitotic marker, phospho-Histone H3 (Fig. 3A) and the mitotic index (Fig. 3B). Ectopic expression of Myc-Mps1 WT, but not Myc-Mps1 KD, at levels comparable to that of the endogenous Mps1 effectively restored mitotic arrest of Mps1 RNAi cells. Our findings confirmed a requirement for the kinase activity of Mps1 in the spindle checkpoint. As compared with Myc-Mps1 WT, Myc-Mps1 T676A had a significantly lower capacity to restore the mitotic arrest in Mps1 RNAi cells upon nocodazole treatment, indicating that Mps1 T676A is deficient in checkpoint signaling (Fig. 3 A and B). On the other hand, the mitotic index and phospho-Histone H3 levels of Myc-Mps1 T676A-expressing cells were higher than those of the Myc-Mps1 KD-expressing cells (Fig. 3 A and B), consistent with the fact that Mps1 T676A had residual kinase activity (Fig. 2A).
Fig. 3.
The Mps1 T676A mutant is defective in spindle-checkpoint function. (A) HeLa tet-on cells were transfected with Mps1 siRNA and the indicated RNAi-resistant Mps1 vectors and then treated with nocodazole. The cell lysates were blotted with the indicated antibodies. The positions of Myc-Mps1 and the endogenous (Endo.) Mps1 are indicated. (B) The mitotic indices of the cells described in A. Approximately 1,000 cells from three independent experiments were counted. The averages and standard deviations are shown. (C) HeLa tet-on cells were transfected with Mps1 siRNA and the indicated RNAi-resistant Mps1 vectors. After incubation with nocodazole for 16 h, the cells were trypsinized, spun down on slides, fixed, and stained with anti-phospho-Histone H3 (red), anti-Myc (green), and DAPI (blue). (D) The mitotic indices of Myc-positive cells described in C. More than 100 cells expressing Myc-Mps1 in two experiments were counted. The average and standard deviations are shown.
Expression of Myc-Mps1 WT did not fully rescue the checkpoint defect of the Mps1 RNAi cells (Fig. 3B). One possible reason might be that the plasmid transfection efficiency was lower than the transfection efficiency of siRNAs. As a result, certain Mps1 RNAi cells were not expressing Myc-Mps1. To circumvent this problem, we stained the transfected cells with anti-Myc, anti-phospho-Histone H3, and DAPI (Fig. 3C). Only Myc-Mps1-positive cells were analyzed. By using this method, ≈70% of Myc-Mps1 WT-expressing cells were arrested in mitosis in the presence of nocodazole, as indicated by their positive phospho-Histone H3 staining (Fig. 3D). In contrast, only ≈35% of the Myc-Mps1 T676A-expressing cells were arrested in mitosis, confirming that the T676A mutation disrupted the spindle checkpoint function of Mps1.
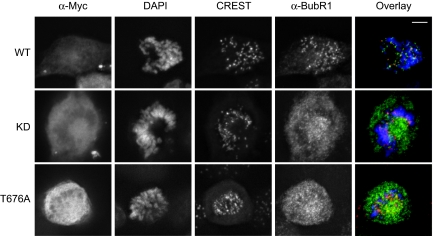
Mps1 is required for the kinetochore localization of other checkpoint proteins, including Mad1, Mad2, and BubR1 (10, 20). We next determined whether Mps1 RNAi cells expressing Myc-Mps1 T676A were defective in the kinetochore localization of BubR1. The ectopically expressed Myc-Mps1 WT, KD, and T676A proteins localized normally to kinetochores in mitosis (SI Fig. 8), suggesting that phosphorylation of T676A is not required for the kinetochore localization of Mps1. Expression of Myc-Mps1 WT restored the kinetochore localization of BubR1 in Mps1 RNAi cells (Fig. 4). In contrast, the vast majority of mitotic Mps1 RNAi cells that expressed Myc-Mps1 KD lacked BubR1 kinetochore staining. A large fraction (12 of 17) of Myc-Mps1 T676A-expressing cells did not exhibit normal BubR1 kinetochore localization. Therefore, phosphorylation of T676A in Mps1 is required for the kinetochore localization of BubR1 and for proper spindle checkpoint signaling.
Fig. 4.
The kinetochore localization of BubR1 is defective in Mps1 RNAi cells expressing Mps1 T676A. HeLa tet-on cells were transfected with Mps1 siRNA and the indicated RNAi-resistant Mps1 vectors. The cells were stained with anti-Myc, CREST (red), anti-BubR1 (green), and DAPI (blue). (Scale bars, 5 μm.)
T676 Phosphorylation of Mps1 Is Regulated During the Cell Cycle.
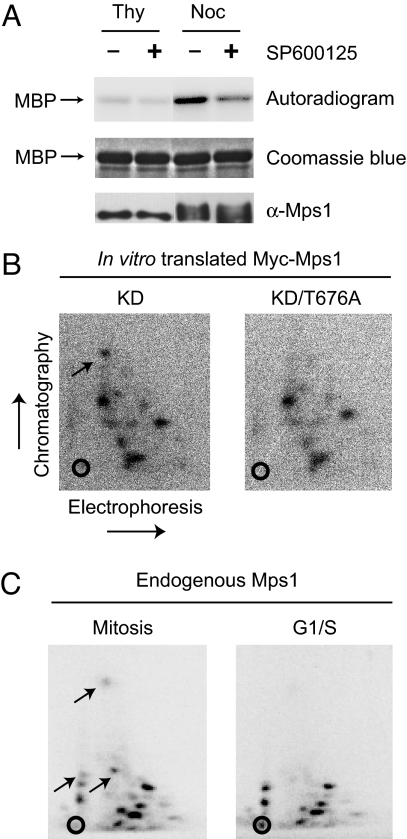
Previous studies have shown that Mps1 is hyperphosphorylated, and its kinase activity is elevated in mitosis (11). To confirm these findings, we IPed Mps1 from thymidine-arrested G1/S HeLa cells or nocodazole-arrested mitotic cells and tested its kinase activity against MBP. Mps1 isolated from mitotic cells migrated more slowly on SDS/PAGE, consistent with it being hyperphosphorylated (Fig. 5A). Furthermore, compared with the IP from G1/S cells, the Mps1 IP from mitotic HeLa cells had higher kinase activity, which was greatly diminished by the addition of a known Mps1 chemical inhibitor, SP600125 (Fig. 5A). Therefore, our results confirm the mitotic activation of Mps1.
Fig. 5.
T676 phosphorylation of Mps1 increases during mitosis. (A) The endogenous Mps1 was IPed from thymidine (Thy)- or nocodazole (Noc)-arrested HeLa tet-on cells and incubated with MBP and γ-[32P]ATP in the presence or absence of the Mps1 inhibitor, SP600125. The reaction mixture was separated on SDS/PAGE and analyzed by autoradiography (Top), Coomassie blue staining (Middle), or anti-Mps1 blotting (Bottom). (B) Phosphopeptide analysis of in vitro translated Myc-Mps1 KD and KD/676A that were phosphorylated by GST-Mps1 in the presence of γ-[32P]ATP. The chromatography and electrophoresis dimensions are labeled. The open circles indicate the sample origin for electrophoresis. The position of the phosphopeptide containing T676 of Mps1 is indicated by an arrow. (C) Phosphopeptide analysis of the endogenous Mps1 that was IPed from G1/S or mitotic HeLa cells metabolically labeled with γ-[32P]-orthophosphate. Phosphopeptides absent in Mps1 from G1/S cells are indicated by arrows.
Using phosphopeptide analysis, we next tested whether phosphorylation of T676 in Mps1 was regulated during the cell cycle. To identify the position of the peptide containing phospho-T676, we first performed phosphopeptide analysis of in vitro translated Myc-Mps1 KD or KD/T676A that had been incubated with GST-Mps1 in the presence of [32P]ATP. The phosphopeptide of Myc-Mps1 KD with the fastest mobility in the chromatography dimension was absent in the Myc-Mps1 KD/T676A sample, indicating that this peptide contained phospho-T676 (Fig. 5B). We then performed phosphopeptide analysis of the endogenous Mps1 isolated from G1/S or mitotic HeLa cells that had been labeled with 32P-phosphate. Although most phosphopeptides were observed in both Mps1 samples, three phosphopeptides were only present in Mps1 from mitotic HeLa cells, but not in Mps1 from G1/S cells (Fig. 5C). One of these mitosis-specific Mps1 phosphopeptides migrated in a position similar to that of the phospho-T676 peptide of the in vitro phosphorylated Mps1. These data suggest that T676 phosphorylation in Mps1 is elevated during mitosis and might be one mechanism responsible for the mitotic activation of Mps1.
Induced Dimerization of Mps1 Is Sufficient for Its Activation.
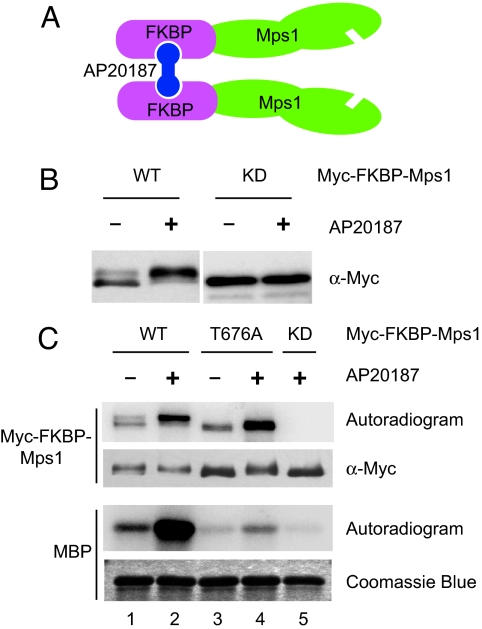
Phosphorylation of residues in the activation loop is a common mechanism for kinase activation (26). Because of the geometry of the active site of kinases, autophosphorylation of their activation loop residues generally occurs in an intermolecular fashion or in trans. Indeed, we have shown above that phosphorylation of Mps1 at T676 can occur through trans autophosphorylation. Therefore, self-association or clustering of Mps1 molecules may enhance T676 phosphorylation and activate its kinase activity. To test this possibility, we fused FK506-binding protein (FKBP) to the N terminus of Mps1. Addition of AP20187, a compound that can bind to two FKBP molecules simultaneously, is expected to induce dimerization of the Myc-FKBP-Mps1 fusion protein (Fig. 6A) (27). HeLa cells were then transfected with plasmids encoding Myc-FKBP-Mps1 WT, KD, and T676A and treated with AP20187. Overexpression of these Mps1 proteins did not significantly increase the mitotic index of HeLa cells in the presence or absence of AP20187 (data not shown). Therefore, unlike in budding yeast, overexpression of Mps1 in human cells is insufficient to cause mitotic arrest in the absence of spindle damage. Addition of AP20187 caused the majority of overexpressed Myc-FKBP-Mps1 WT, but not KD, to exhibit slower gel mobility (Fig. 6B). Furthermore, Myc-FKBP-Mps1 WT from cells treated with AP20187 had much higher kinase activity toward MBP than did Mps1 from untreated cells (Fig. 6C). AP20187 treatment failed to activate the kinase activity of Myc-FKBP-Mps1 T676A. These results suggest that induced dimerization of Mps1 is sufficient to promote Mps1 activation in cells through autophosphorylation at T676.
Fig. 6.
Induced dimerization of Mps1 increases its kinase activity in cells. (A) Schematic drawing of the FKBP-Mps1 fusion protein and its chemical-induced dimerization. (B) HeLa tet-on cells were transfected with plasmids encoding Myc-FKBP-Mps1 WT or KD and incubated with or without AP20187. The total cell lysate was blotted with anti-Myc. (C) The Myc-FKBP-Mps1 proteins were IPed from lysates of cells described in B and assayed for their kinase activity against MBP in the presence of γ-[32P]ATP. The reaction mixtures were separated on SDS/PAGE and analyzed by autoradiography, anti-Myc blotting, or Coomassie blue staining.
As expected, induced dimerization of Mps1 also enhanced the autophosphorylation of FKBP-Mps1 WT (Fig. 6C; compare lanes 1 and 2). Surprisingly, the FKBP-Mps1 T676A mutant still underwent autophosphorylation, which was further enhanced by AP20187 (Fig. 6C; compare lanes 3 and 4). This surprising finding suggests that, even though T676 is a major autophosphorylation site within the kinase domain of Mps1 (Fig. 2C), the full-length Mps1 contains additional autophosphorylation sites. This finding further suggests that the T676A mutation might selectively decrease the interaction between Mps1 and its substrates (i.e., increase Km) without significantly decreasing the catalytic rate constant (kcat) of Mps1. Thus, Mps1 T676A might be less active because it does not interact efficiently with substrates. Induced dimerization of FKBP-Mps1 T676A raises the local substrate concentration for Mps1 and might overcome the effect of T676A mutation in increasing Km of the autophosphorylation reaction.
Discussion
Requirement for T676 Phosphorylation in Mps1 Activation.
In this study, we have shown that phosphorylation at T676, a residue located in the activation loop of Mps1, is both required for in vitro kinase activity of Mps1 and for its checkpoint function in cells. Phosphorylation of activation loop residues is a common mechanism for the activation of protein kinases (26). In most cases, the phosphate group in the phosphorylated activation loop engages in favorable electrostatic interactions with the arginine (R) preceding the catalytic aspartate (D) residue in the so-called RD kinases (26). These interactions help to organize the activation loop into an active conformation that is more suitable for substrate-binding and catalysis. Mps1, however, is not an RD kinase (SI Fig. 7). So, how does T676 phosphorylation activate Mps1? Because of the lack of the RD pocket in Mps1, other basic residues in Mps1 might interact with the phosphate on T676 to stabilize the active conformation of its activation loop. Along this vein, Mps1 contains a cluster of basic residues that are located C-terminal to its activation segment, including K706, K708, and K710 (SI Fig. 7). Based on our structure model of the Mps1 kinase domain, these basic residues are in close proximity to T676 and can potentially serve as binding partners for phospho-T676 (Fig. 1D). Consistently, mutation of K708 and K710 to alanines greatly reduced the kinase activity of Mps1 (data not shown).
In addition to its C-terminal kinase domain, Mps1 contains an N-terminal tetratricopeptide repeat (TPR) domain, which is involved in its kinetochore localization (12, 28, 29). Aside from T676, we have identified nine other phosphorylation sites on Mps1. S821 is located just C-terminal to the kinase domain of Mps1. The corresponding residue, S844, in Xenopus Mps1 has been shown to be phosphorylated, and this phosphorylation event is required for the kinetochore targeting of Mps1 in Xenopus egg extracts (24). Recently, Kasbek et al. showed that cyclin A/Cdk2 phosphorylates an Mps1 fragment at T468 in vitro (25). The Mps1 T468A mutant failed to accumulate at centrosomes, suggesting that this phosphorylation event antagonizes centrosome-specific degradation of Mps1 (25). Other phosphorylation sites of Mps1 are located in the central regions between its TPR and kinase domains. The functions of these phosphorylation events remain to be elucidated.
Autophosphorylation-Dependent Activation of Mps1 in Mitosis.
The ligand-induced dimerization of certain receptor tyrosine kinases enhances their trans autophosphorylation at a tyrosine residue in the activation loop, leading to the activation of their kinase activity (30). Recently, it has been proposed that the enrichment of Aurora B-containing kinase complex at chromosomes leads to its autoactivation (31). The kinase activity of Mps1 is required for the spindle checkpoint and is activated during mitosis (11). Can a similar mechanism be responsible for Mps1 activation in mitosis? We have shown that phosphorylation of T676 in Mps1 increases during mitosis, suggesting that it might be a mechanism for the mitotic activation of Mps1. Although an upstream kinase may also contribute to this phosphorylation in vivo, phosphorylation of T676 in Mps1 can occur through trans autophosphorylation. Thus, mechanisms that promote the clustering or self-association of Mps1 are expected to enhance the trans autophosphorylation at T676, thereby activating Mps1. The protein levels of Mps1 are higher in mitosis. In addition, Mps1 is recruited to kinetochores during mitosis. These two factors are expected to raise the local concentration of Mps1 at kinetochores. The clustering of Mps1 at kinetochores may lead to more efficient trans autophosphorylation of Mps1 at T676 and possibly other positions, resulting in the activation of Mps1 SI Fig. 9).
As a first step toward testing this model, we have shown that induced dimerization of Mps1 is indeed sufficient to activate Mps1 in cells. We have, however, failed to detect significant self-association between ectopically expressed, differentially tagged Mps1 molecules in interphase or mitotic HeLa cell lysates (data not shown). Thus, oligomerization of Mps1 might only occur at kinetochores or might be bridged through other cellular proteins.
In summary, Mps1 undergoes trans autophosphorylation of an activation-loop residue in mitosis, which is required for the activation of Mps1 and for the spindle checkpoint. It remains to be determined whether this phosphorylation event is an initiating step of spindle checkpoint signaling at kinetochores or a part of a positive feedback loop that amplifies the checkpoint signal.
Materials and Methods
Plasmids and siRNAs.
Full-length human Mps1 was isolated from human fetal thymus cDNA (Clontech) and cloned into the appropriate vectors. The Mps1 mutants were generated by using the QuikChange Site-directed Mutagenesis Kit (Stratagene). The FKBP-coding region was amplified from pC4M-FV2E (ARIAD) by PCR and cloned into the pCS2-Myc-Mps1 vector. siRNA oligonucleotides of MPS1 (5′-UGAACAAAGUGAGAGACAU-3′) were synthesized by Dharmacon. siRNA-resistant Mps1 plasmids was created by introducing silent mutations in the siRNA-targeting region.
Cell Culture, Transfections, and Metabolic Labeling.
HeLa tet-on (Clontech) and HeLa S3 cells were grown in DMEM (Invitrogen) supplemented with 10% FBS. Transfections of siRNAs and plasmids were performed with Lipofectamine RNAi MAX (Invitrogen) and Effectene (Qiagen), respectively, according to manufacturers' instructions. Metabolic labeling of HeLa tet-on cells was performed as described in ref. 32. To induce dimerization of Myc-FKBP-Mps1, cells transfected with the appropriated plasmids were incubated with 10 nM AP20187 (ARIAD).
Antibodies and Immunoprecipitation (IP).
Rabbit polyclonal antibodies against human Mps1 were generated by Zymed by using a GST-fusion protein containing the N-terminal 200 residues of Mps1. Production of antibodies against human BubR1 was described in ref. 33. The following antibodies were purchased from the indicated commercial sources: CREST antibody (ImmunoVision), anti-Myc antibody (Roche), and anti-phospho-histone H3 antibody (Upstate Biotechnology). Large-scale IP of Mps1 and immunofluorescence were performed as described in ref. 34.
Mass Spectrometry.
The protein band was excised and destained. Gel pieces were digested with trypsin (Promega). The tryptic peptides were analyzed on a LTQ 2-D ion trap mass spectrometer (ThermoFisher) coupled with an Agilent 1100 nano-flow HPLC system (Agilent) with a reversed-phase C18 capillary column. Tandem mass spectra of the peptides were acquired in the data-dependant mode and searched against NCBI-nr database with Mascot software (v2.1). All results were manually verified.
Phosphopeptide Mapping.
Phosphopeptide mapping was performed with the HTLE system (CBS Scientific) according to the manufacturer's instructions.
In Vitro Kinase Assays.
For IP-kinase assays, the Myc IPs were incubated with MBP in kinase buffer I [50 mM salt Tris (pH 7.5), 0.15 M NaCl, 10 mM MgCl2, 1 mM DTT, 5 mM NaF, 20 mM β-glycerophosphate, 0.1 mM Na3VO4] containing 0.1 mM ATP and 0.1 μCi/ μl γ-[32P]ATP for 30 min at 30°C. The reaction mixture was quenched with SDS sample buffer, separated on SDS/PAGE, and analyzed by a phosphoimager. For inhibition of Mps1, 2 μM of SP600126 (Biomol) was added in the kinase reactions. For the Mps1 reactivation experiment, GST-Mps1 was IPed by using anti-Mps1 beads from Sf9 cells lysed in kinase buffer II [50 mM Tris (pH 7.5), 0.2 M NaCl, 10 mM MgCl2, 1 mM DTT], treated with λ phosphatase (NEB) for 30 min at 30°C, and incubated in kinase buffer II containing 0.2 mM ATP for 30 min at 30°C. The samples were then incubated with MBP in kinase buffer II containing 0.2 mM ATP and 0.1 μCi (1 Ci = 37 GBq)/μl γ-[32P]ATP for 30 min at room temperature.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Dr. Yan Li (Protein Chemistry and Technology Center, University of Texas Southwestern) for initial mass spectrometry analysis of Mps1. This work is supported by the National Institutes of Health (GM61542), the W. M. Keck Foundation, the March of Dimes Foundation, the Welch Foundation, and the Leukemia and Lymphoma Society.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710519105/DC1.
References
- 1.Kops GJ, Weaver BA, Cleveland DW. Nat Rev Cancer. 2005;5:773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- 2.Bharadwaj R, Yu H. Oncogene. 2004;23:2016–2027. doi: 10.1038/sj.onc.1207374. [DOI] [PubMed] [Google Scholar]
- 3.Musacchio A, Salmon ED. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 4.Yu H. Mol Cell. 2007;27:3–16. doi: 10.1016/j.molcel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 5.Raman M, Chen W, Cobb MH. Oncogene. 2007;26:3100–3112. doi: 10.1038/sj.onc.1210392. [DOI] [PubMed] [Google Scholar]
- 6.Winey M, Goetsch L, Baum P, Byers B. J Cell Biol. 1991;114:745–754. doi: 10.1083/jcb.114.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss E, Winey M. J Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- 9.Fisk HA, Winey M. Cell. 2001;106:95–104. doi: 10.1016/s0092-8674(01)00411-1. [DOI] [PubMed] [Google Scholar]
- 10.Abrieu A, Magnaghi-Jaulin L, Kahana JA, Peter M, Castro A, Vigneron S, Lorca T, Cleveland DW, Labbe JC. Cell. 2001;106:83–93. doi: 10.1016/s0092-8674(01)00410-x. [DOI] [PubMed] [Google Scholar]
- 11.Stucke VM, Sillje HH, Arnaud L, Nigg EA. EMBO J. 2002;21:1723–1732. doi: 10.1093/emboj/21.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu ST, Chan GK, Hittle JC, Fujii G, Lees E, Yen TJ. Mol Biol Cell. 2003;14:1638–1651. doi: 10.1091/mbc.02-05-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poss KD, Nechiporuk A, Hillam AM, Johnson SL, Keating MT. Development (Cambridge, UK) 2002;129:5141–5149. doi: 10.1242/dev.129.22.5141. [DOI] [PubMed] [Google Scholar]
- 14.Poss KD, Wilson LG, Keating MT. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 15.Fischer MG, Heeger S, Hacker U, Lehner CF. Curr Biol. 2004;14:2019–2024. doi: 10.1016/j.cub.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 16.Gilliland WD, Wayson SM, Hawley RS. Curr Biol. 2005;15:672–677. doi: 10.1016/j.cub.2005.02.062. [DOI] [PubMed] [Google Scholar]
- 17.Gilliland WD, Hughes SE, Cotitta JL, Takeo S, Xiang Y, Hawley RS. PLoS Genet. 2007;3:e113. doi: 10.1371/journal.pgen.0030113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei JH, Chou YF, Ou YH, Yeh YH, Tyan SW, Sun TP, Shen CY, Shieh SY. J Biol Chem. 2005;280:7748–7757. doi: 10.1074/jbc.M410152200. [DOI] [PubMed] [Google Scholar]
- 19.Zhu S, Wang W, Clarke DC, Liu X. J Biol Chem. 2007;282:18327–18338. doi: 10.1074/jbc.M700636200. [DOI] [PubMed] [Google Scholar]
- 20.Vigneron S, Prieto S, Bernis C, Labbe JC, Castro A, Lorca T. Mol Biol Cell. 2004;15:4584–4596. doi: 10.1091/mbc.E04-01-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones MH, Huneycutt BJ, Pearson CG, Zhang C, Morgan G, Shokat K, Bloom K, Winey M. Curr Biol. 2005;15:160–165. doi: 10.1016/j.cub.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt M, Budirahardja Y, Klompmaker R, Medema RH. EMBO Rep. 2005;6:866–872. doi: 10.1038/sj.embor.7400483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mattison CP, Old WM, Steiner E, Huneycutt BJ, Resing KA, Ahn NG, Winey M. J Biol Chem. 2007;282:30553–30561. doi: 10.1074/jbc.M707063200. [DOI] [PubMed] [Google Scholar]
- 24.Zhao Y, Chen RH. Curr Biol. 2006;16:1764–1769. doi: 10.1016/j.cub.2006.07.058. [DOI] [PubMed] [Google Scholar]
- 25.Kasbek C, Yang CH, Mohd Yusof A, Chapman HM, Winey M, Fisk HA. Mol Biol Cell. 2007;18:4457–4469. doi: 10.1091/mbc.E07-03-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nolen B, Taylor S, Ghosh G. Mol Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 27.Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 28.Yu H. Curr Opin Cell Biol. 2002;14:706–714. doi: 10.1016/s0955-0674(02)00382-4. [DOI] [PubMed] [Google Scholar]
- 29.Stucke VM, Baumann C, Nigg EA. Chromosoma. 2004;113:1–15. doi: 10.1007/s00412-004-0288-2. [DOI] [PubMed] [Google Scholar]
- 30.Schlessinger J. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 31.Kelly AE, Sampath SC, Maniar TA, Woo EM, Chait BT, Funabiki H. Dev Cell. 2007;12:31–43. doi: 10.1016/j.devcel.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sefton BM, Beemon K, Hunter T. J Virol. 1978;28:957–971. doi: 10.1128/jvi.28.3.957-971.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang Z, Bharadwaj R, Li B, Yu H. Dev Cell. 2001;1:227–237. doi: 10.1016/s1534-5807(01)00019-3. [DOI] [PubMed] [Google Scholar]
- 34.Tang Z, Shu H, Qi W, Mahmood NA, Mumby MC, Yu H. Dev Cell. 2006;10:575–585. doi: 10.1016/j.devcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.