Abstract
The chemical sense of insects has evolved to encode and classify odorants. Thus, the neural circuits in their olfactory system are likely to implement an efficient method for coding, processing, and classifying chemical information. Here, we describe a computational method to process molecular representations and classify molecules. The three-step approach mimics neurocomputational principles observed in olfactory systems. In the first step, the original stimulus space is sampled by “virtual receptors,” which are chemotopically arranged by a self-organizing map. In the second step, the signals from the virtual receptors are decorrelated via correlation-based lateral inhibition. Finally, in the third step, olfactory scent perception is modeled by a machine learning classifier. We found that signal decorrelation during the second stage significantly increases the accuracy of odorant classification. Moreover, our results suggest that the proposed signal transform is capable of dimensionality reduction and is more robust against overdetermined representations than principal component scores. Our olfaction-inspired method was successfully applied to predicting bioactivities of pharmaceutically active compounds with high accuracy. It represents a way to efficiently connect chemical structure with biological activity space.
Keywords: bioinformatics, chemical biology, computational model, decorrelation, olfactory coding
The mechanisms that enable olfactory discrimination are remarkably similar across species and even phyla (1, 2), and several principles of organization have been observed in insects and vertebrates. One such principle is that each primary olfactory sensory neuron (OSN) specifically expresses one type of olfactory receptor (OR), as has been demonstrated, e.g., in mice (3, 4) and in Drosophila (5, 6), although exceptions to this rule exist (7, 8). ORs represent the largest family of seven-transmembrane G protein-coupled receptors (9–12). Several studies addressed structure–activity relationships (SARs) of ORs (13–19). A general observation is that one odorant typically activates a number of different ORs, and each OR has rather broad ligand selectivity. Investigation of Drosophila OR neurons (20) also indicated that each receptor preferably responds to a specific combination of chemical features; that is, each receptor samples a specific region of “chemical space.”
Another characteristic of olfactory systems is that OSNs expressing a specific OR make synaptic contacts with a defined subset of second-order neurons in downstream neural populations, namely the olfactory bulb of vertebrates (21) or the antennal lobe of insects (22, 23). These connections are formed in spatially discrete areas, the glomeruli. It has long been speculated, and in part also shown, that glomeruli are chemotopically ordered, such that neighboring glomeruli receive input from OSNs that prefer ligands with similar chemical characteristics (5, 24–27). There is some evidence that the spatial distance of glomeruli in the olfactory bulb is related to the distance between their genomic sequences (5). It has also been demonstrated that receptor sequence similarity at least in some cases correlates with the chemical properties of preferred ligands (28–30). Such a chemotopic organization in the secondary processing stage of olfactory information can be exploited for computational processes to decorrelate input signals (31). In particular, correlation-based lateral inhibition can explain the processing of receptor activation patterns in the antennal lobe (32).
Neurons extrinsic to the secondary structure project to overlapping regions in brain areas that receive input from all sensory modalities (33–37). All information necessary to assign a perceptual quality to a chemical stimulus is present there.
In summary, insect and vertebrate olfactory systems can be subdivided into three stages of functional organization. In the first stage, OSNs encode the stimulus features into neuronal signals. The second stage decorrelates these signals. In the third stage, the processed representations (“patterns”) are associated with perceptual qualities. Regarding these parallels in organization of neural connectivity, the question arises as to whether this architecture has properties that make it superior to other coding strategies for chemical information. Here, we present a general computational model that processes chemical information following this three-steps design. We show that signal decorrelation in the second stage of olfactory signal processing facilitates odorant classification, which might be relevant for the perception of scent qualities.
Results
In the olfactory system, chemical information is translated into neuronal signals, which undergo processing as they are relayed to higher brain centers. We wondered whether the processing principles in the olfactory systems can generally be applied to information processing and enhance our ability to classify chemical data. To address this issue, we designed a simplified computational model of the three basic processing stages in the olfactory system.
Step 1: Modeling Response Patterns.
In the first step of the model, odorant stimuli were encoded by using “virtual receptors.” Just as real ORs respond to ligands sharing similar properties, a virtual receptor will respond to ligands that occupy the same region in chemical space. Fig. 1A illustrates the concept of the virtual receptor: The smaller the distance between an odorant and the virtual receptor in descriptor space, the higher the response (or activation) of this virtual receptor will be. For our analysis, 836 odorants were taken from the Sigma–Aldrich Flavors and Fragrances (38). Odorants were represented by 184 molecular descriptors. Hence, each odorant is described by a 184-dimensional vector, each element of which encodes a chemical property, forming the basis of a 184-dimensional “chemical space.”
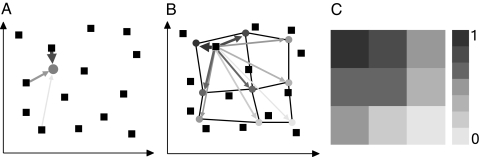
Fig. 1.
Creation of virtual response patterns, schematic. (A) Arrow weight and shading depict the degree of activation of a virtual receptor (gray disk) by odorants (squares). (B) Activation of a grid of virtual receptors by one odorant. Lines connecting receptors symbolize neighborhood relationships in the SOM topology. (C) The resulting response pattern. Each rectangle corresponds to one receptor, where shades of gray indicate the strength of activation, as depicted on the scale to the right.
Considering an array of n virtual receptors, each receptor has a position described by a coordinate vector p in the m-dimensional descriptor space. The response of a virtual receptor to an odorant should increase with decreasing distance between an odorant and the receptor in chemical descriptor space. We defined the response ri of the ith receptor (i = 1, 2, … n) to an odorant, s, as
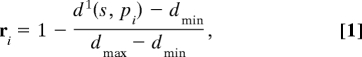 |
where pi represents the coordinates of the ith receptor, d1(s, pi) is the Manhattan distance (Minkowski metric with k = 1, the sum of absolute coordinate differences) between s and pi, and dmin and dmax are the minimal and maximal distance between any s and pi. Thus, ri = 0 if d1(s, pi) is maximal and ri = 1 if d1(s, pi) is minimal.
The coordinates of the receptors should be chosen such that they cover all relevant parts of chemical space. We used a self-organizing map (SOM) to arrange our virtual receptors in this data space (39). SOMs are capable of preserving local topology in their low-dimensional projections (40). This neighborhood-preserving organization naturally leads to a chemotopic arrangement such that neighboring units are more similar in their ligand characteristics than units that are more separated on the SOM (Fig. 1B). The activity pattern can be arranged on a two-dimensional plane according to the projection that is defined by the SOM's topology (Fig. 1C). Each rectangle corresponds to the output of one virtual receptor, shades of gray indicate different degrees of their activation by the odorant stimulus.
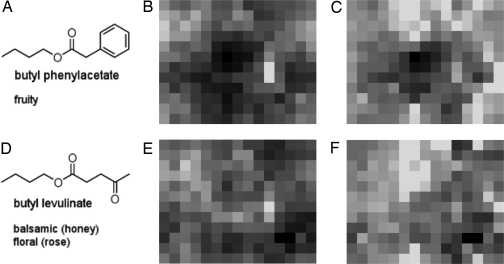
The SOMs we trained had toroidal architecture and thus can be visualized as two-dimensional grids. In the scope of our model, a trained SOM corresponds to the arrangement of glomeruli in the antennal lobe. Fig. 2 depicts two odorants (Fig. 2A, butyl phenylacetate; Fig. 2D, butyl levulinate) and the respective response patterns (Fig. 2 B and E) from an SOM with 12 × 15 virtual receptors. Due to the toroidal grids, the upper and lower edges of the patterns are connected, as are the left and right edges.
Fig. 2.
Virtual response patterns for two odorants before and after processing. (A and D) Odorants and their scent qualities. (B and E) Virtual response patterns evoked by the odorants. (C and F) Processed response patterns. Black corresponds to maximal activation, and light gray corresponds to minimal activation.
Within the scope of the model, the evoked response patterns correspond to activation of glomeruli in the antennal lobe. The patterns generated by our model are similar to those observed in animals: Each odorant activates multiple receptors (glomeruli), and each receptor is activated by several odorants. Most of the patterns showed multiple “islands” of high activation; thus, most odorants activated several units that are not necessarily neighbors on the SOM grid. These activation islands reflect that the SOM corresponds to a manifold rather than a hyperplane in the descriptor space; i.e., it is “folded” and not planar. In part, the curvature is due to the toroidal structure of the SOM but may also be a consequence of the neighborhood structure in odorant space.
In summary, the virtual response pattern adequately mirrors real receptor activation by odorants and their downstream targets, that is, the glomeruli. The patterns represent the raw signal evoked by a chemical structure. The next step of olfactory information processing is decorrelation of these receptor responses.
Step 2: Pattern Processing by Lateral Inhibition.
It has been proposed that processing in the antennal lobe implements correlation-based lateral inhibition (32). If two glomeruli respond similarly to a set of ligands, they will inhibit each other's response. This enforces a “winner takes most” situation such that the glomerulus with the stronger response will inhibit the response of the weaker glomerulus, effectively making their output more dissimilar. The more correlated the firing patterns of the two glomeruli, the more pronounced this effect will be.
To account for lobal processing, we computed the postlobal pattern vector r′ from the prelobal input vector r (compare with Eq. 1) by Eq. 2
where n is the number of virtual receptors, q is an arbitrary weight, and C is a matrix, with Ci,j containing the Pearson correlation coefficient for the responses of the ith and jth receptor. Negative elements and all elements on the diagonal of C were set to zero.
Fig. 2 C and F shows computed postlobal response patterns. The most salient difference between the patterns is that there is less overall activation. Notably, the sites of highest activation remain unchanged (in the center in Fig. 2C and in the lower left in Fig. 2F), whereas large portions of the remaining pattern get sparser (i.e., show less activity).
In summary, lateral inhibition between correlated units results in more focused signals. The decorrelated response patterns form the basis for subsequent scent perception.
Step 3: Pattern Classification by Machine Learning.
The third step of olfactory information processing, the assignment of a perceptual quality, was modeled as a machine learning process. We performed a retrospective scent prediction experiment to examine the impact of the above processing stages on the accuracy of scent quality prediction.
We used odor annotations to 836 odorants from the 2004 Sigma–Aldrich Flavors and Fragrances (38) as targets to train a Naive–Bayes classifier, as implemented in the WEKA machine learning suite (41). We chose this particular method because it has no tunable parameters and, thus, is optimally suited for classification without the need to optimize additional free variables. In this study, the Naive–Bayes approach served the sole purpose of providing a parameter-free classifier for measuring the relative effect of the first two steps of our olfactory information processing model.
After removing scents that occurred less than five times in the data set, we obtained a total of 66 scent qualities. Each odorant was allowed to have several scent annotations. We trained the classifier separately for each scent class; hence, each of the 66 resulting classifiers only distinguished between, e.g., “smoky” and “not smoky” or “fruity (Banana)” and “not fruity (Banana)” of the 836 compounds bearing this attribute. We assessed prediction performance by the area under the receiver–operating characteristic curve [area under curve (AUC)] (41). A classifier was trained 50 times by using 5-fold cross-validation (80 + 20 data split of training and test data), obtaining 50 AUC values per scent quality, of which we formed the median.
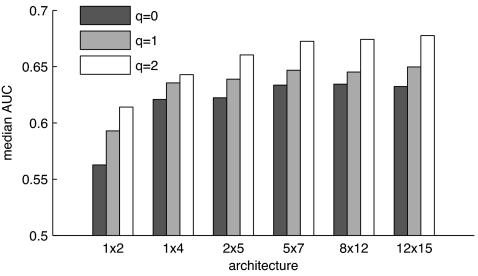
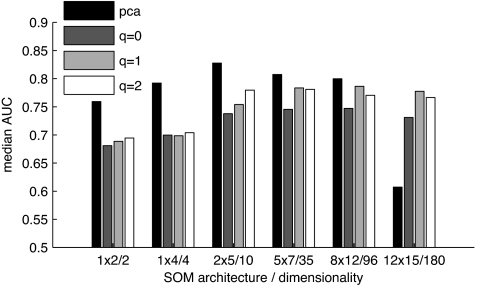
Our three-steps approach has only two free parameters that both affected classification performance: the SOM size (i.e., number of virtual receptors) and q (the weight of correlational inhibition; compare with Eq. 2). To analyze their effect, we trained SOMs containing between 2 and 180 virtual receptors (abscissa in Fig. 3) and used different values of q to process the activation patterns. Results are depicted in Fig. 3: Patterns generated with q = 2 outperformed q = 0 (no processing) and q = 1 for almost all SOM architectures. For the 12 × 15 representations, the median AUC values were 0.68 (q = 2), 0.65 (q = 1), and 0.63 (q = 0). These differences are significant (P <10−7; Wilcoxon rank sum test). Performance also gradually decreased with dimensionality, but only 2 × 5 and smaller representations significantly differ from the larger representations in their median AUC values (P < 0.05; Wilcoxon rank sum test). Hence, overall classification performance did not suffer from a reduction of dimensionality by a factor of five. For comparison, we also tested the Naive–Bayes classifier on the initial 184 chemical descriptors without processing (except scaling and centering) and yielded a median AUC of 0.67. This value indicates that the olfaction-inspired decorrelation step does not lead to a loss of information but can even help to improve classification.
Fig. 3.
Impact of SOM architecture and correlational processing on classification performance. Median AUC values for scent prediction using unprocessed patterns (q = 0) and patterns processed by correlational inhibition, with q = 1 and q = 2. The ordinate was truncated to emphasize differences.
The Influence of Signal Decorrelation on Classification.
One hypothesis for the functional meaning of processing in the antennal lobe is that it decorrelates receptor signals (42). In the present work, this decorrelation is achieved through mutual inhibition between projection neurons that innervate glomeruli with correlated response patterns. To quantify the amount of decorrelation, we calculated the residual correlation between virtual receptors for various settings of q.
Although in the unprocessed receptor responses there was high residual correlation (q = 0; mean correlation = 0.38), it gradually decreased with increasing q; for q = 1 the mean residual correlation is 0.24, whereas it decreased to 0.02 for q = 2. Also for q = 2, there is some residual correlation, although its values are distributed around zero, hence the small mean correlation. In contrast, the dimensions produced by principal component analysis (PCA) are orthogonal and have zero residual correlation. PCA is frequently used for dimensionality reduction before training machine learning classifiers (43).
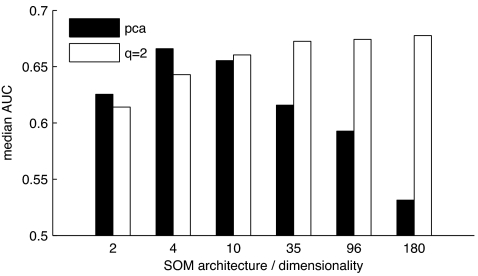
PCA by definition achieves maximum decorrelation of the data, but it is not clear beforehand if the resulting patterns also are better suited for classification. To investigate whether maximum decorrelation also corresponds to maximal classification performance, we compared the classification performance of our method with the performance that can be achieved by using PCA for dimensionality reduction on the original descriptor set. Fig. 4 shows median AUC values from retrospective classification using patterns processed with q = 2 and using the first n principal components of the original data set that explained most variance. We chose the dimensionality of the reduced data to match the dimensionality of the patterns.
Fig. 4.
Classification performance using patterns processed by correlation-based inhibition vs. PCA scores of the original descriptors.
The patterns filtered by correlation-based lateral inhibition yielded higher AUC values for higher dimensionalities (maximum = 0.68 by using 180 dimensions), whereas the principal components seemed to work best for low dimensionalities (maximum = 0.67 by using four dimensions). A possible explanation for this behavior is that, with PCA, the dimensions explaining less variance introduce noise, leading to inaccurate training and decreased performance of the classifier.
Therefore, PCA may be a method of choice for data preprocessing if dimensionality reduction is important, e.g., if computational resources are limited, but care must be taken not to use too many principal components for data representation. Due to their highly parallel architecture of “real” brains, data dimensionality may not be the limiting factor. Rather, robustness to noise and capacity of the code may be more important. The latter is provided by the higher dimensionality of the proposed coding scheme, whereas robustness can be increased by the residual redundancy due to nonabsolute decorrelation.
Prediction of the Pharmacological Activity of Compounds.
We then tested whether our method also is suited for pharmaceutical data. This time, chemical space was given by the COBRA database (version 6.1) containing pharmacologically active substances (44). We used the same processing scheme as for the odorant data. Classifiers were trained on the activation patterns, and their “perceptual qualities” were given by the annotated activity at 115 pharmaceutical targets (e.g., cyclooxygenase 2 or thrombin) and their superclasses (e.g., enzyme, G protein coupled receptor, or ion channel). We repeated cross-validation only 10 times (in contrast to 50 times in the previous experiment) to save computing time. We also trained classifiers on the original descriptors processed by PCA. Fig. 5 shows the median AUC values obtained for the pharmaceutical data.
Fig. 5.
Classification performance for pharmaceutical activity data.
One point of difference to the results above is that overall performance was better on the pharmaceutical data set. For example, the 2 × 5 architecture with q = 2 yielded a median AUC value of 0.78 on the drug data set, in contrast to 0.66 on the odorant set. In addition, 49 of 115 targets yielded AUC values of >0.8, indicating that our method is indeed suited for prediction of biological activity.
Another difference to the odorant set is in the performance for high dimensionalities. Although the virtual response patterns outperformed principal components for the highest dimensionality, this trend is less obvious than in the odorant data set. Still, when retaining maximum dimensionality, the virtual response patterns perform best.
A third point of difference is that patterns transformed with q = 2 were not always performing best. Analysis of the filtered patterns revealed that, for q = 2 and dimensionalities higher than 2 × 5, many virtual receptor responses vanished because the subtractive term in Eq. 2 became equal or greater than ri (data not shown). Thus, only the virtual receptor signals with highest activation “survived” lateral inhibition, effectively replacing the soft winner-takes-most situation by a hard winner-takes-all one, with an overall negative impact on classification. This result points out the need for q to be adjusted to obtain optimal results. It also shows that there is not one optimal setting of q but rather that this optimum depends on the input data.
Discussion
We have presented a computational framework that implements processing principles observed in olfactory systems. This method effectively captured relevant properties of the original data that allowed a machine learning classifier to learn odorant classification. Besides reducing dimensionality of the original data, it also exhibited robustness against overdetermined representations, a situation where principal components of the original data failed. In addition, the application of this framework is not limited to the olfactory domain but can also be efficiently used for virtual screening of a pharmaceutical compound database. The results may be further improved if optimal model parameters are chosen and more advanced machine learning systems are used. We see primary applications of our decorrelation technique for low-dimensional mapping of complex data manifolds (45), for example, in SAR modeling and virtual compound screening, especially in a highly parallel context.
The processing scheme we present here provides a simplified model of neural computation in the olfactory system. Our focus was on providing a framework that enables us to study certain aspects of computational principles, instead of trying to build a biologically accurate simulation of the olfactory system. We tried to keep the simulation overhead as small as possible so that the essence of the processing strategies would stay obvious. More realistic models in terms of biological plausibility are particularly useful when one tries to answer biological questions (42, 46, 47). Consequently, we use the prediction performance as a relative measure to compare different mechanisms for processing in the antennal lobe. The results should not be interpreted as providing an actual prediction method for scent.
The q factor showed to have a large impact on the outcome of the correlational filtering step, with classifiability of the patterns improving for rising values of q, up to a certain level. Finding optimal values of q for a given data set may be a worthwhile topic for further research. Possible approaches include the use of metaoptimization techniques to derive q empirically, as demonstrated for the number of hidden neurons in an artificial neural network (48), or its estimation from statistical properties of the data, like variance or cross-correlation.
Among the questions we did not address here are the effects of odorant concentration and combinations of odorants (mixtures). For both questions, we can suggest straightforward implementations: Odorant concentration could be implemented via “gain control” of the activation patterns, i.e., multiplication of the pattern with a concentration-dependent scalar, whereas odorant mixtures could be represented by additive or even nonlinear combinations of their activation patterns. The effects of processing in the virtual antennal lobe on those extensions as well as their impact on classification power provide a tantalizing prospect for future research.
Methods and Data
Source Data.
The chemical space for this experiment was defined by a set of 836 odorants from ref. 38, with scent qualities obtained from the “organoleptic properties” section of the catalog.
Descriptor Calculation.
Molecules and their odor components were extracted from the Sigma–Aldrich Flavors and Fragrances catalog (38). Using their accession numbers, all compounds were carefully checked for correctness with the machine-readable form of the Sigma catalog. Three-dimensional molecular models were obtained with CORINA (Molecular Networks) by using one conformer per molecule. Partial charges were computed with MOE Version 2005.06 (Chemical Computing Group) by using the MMFF94x force field [a modified version of MMFF94s (49)].
Before descriptor calculation, we performed an additional energy minimization by using MOE and the MMFF94x force field, stopping at a gradient of 10−4. Descriptors were calculated by using MOE. We used the complete set of available 2D descriptors, resulting in a 184-dimensional descriptor space. Although we used only 2D descriptors, we calculated the 3D models because a molecule's conformation affects the distribution of partial charges, which is relevant for some 2D descriptors.
SOM Training.
We used SOMMER for SOM training (39). Molecular descriptors were scaled to unit variance and zero mean before SOM training. We used the Manhattan distance function, tmax = 70,000, σi = 5.0, σf = 0.1, λi = 0.7, and λf = 0.01 for SOM training for all variants except the 1 × 2 SOM, for which we used tmax = 100 and σf = 0.5. Descriptor vectors were presented in random sequence.
Machine Learning and Performance Assessment.
We used the Naive–Bayes classifier as implemented in the WEKA machine learning suite (41) for all classification experiments. Probabilities were estimated assuming a normal distribution for the feature vectors. ROC curves were generated by arranging compounds by decreasing predicted probability of class adherence and cumulatively calculating rates of false and true positives.
ACKNOWLEDGMENTS.
We thank Volker Majczan for assistance in the preparation of the odorant database and Natalie Jäger and Joanna Wisniewska for preliminary experiments on odorant classification that were helpful in the design of this study. This work was supported by the Beilstein-Institut zur Förderung der Chemischen Wissenschaften.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Hildebrand JG, Shepherd GM. Annu Rev Neurosci. 1997;20:595–631. doi: 10.1146/annurev.neuro.20.1.595. [DOI] [PubMed] [Google Scholar]
- 2.Firestein S. Nature. 2001;413:211–218. doi: 10.1038/35093026. [DOI] [PubMed] [Google Scholar]
- 3.Chess A, Simon I, Cedar H, Axel R. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- 4.Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 5.Couto A, Alenius M, Dickson BJ. Curr Biol. 2005;15:1535–1547. doi: 10.1016/j.cub.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 6.Fishilevich E, Vosshall LB. Curr Biol. 2005;15:1548–1553. doi: 10.1016/j.cub.2005.07.066. [DOI] [PubMed] [Google Scholar]
- 7.Mombaerts P. Curr Opin Neurobiol. 2004;14:31–36. doi: 10.1016/j.conb.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 8.Goldman AL, der Goes van Naters WV, Lessing D, Warr CG, Carlson JR. Neuron. 2005;45:661–666. doi: 10.1016/j.neuron.2005.01.025. [DOI] [PubMed] [Google Scholar]
- 9.Buck L, Axel R. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 10.Mombaerts P. Science. 1999;286:707–711. doi: 10.1126/science.286.5440.707. [DOI] [PubMed] [Google Scholar]
- 11.Gao Q, Chess A. Genomics. 1999;60:31–39. doi: 10.1006/geno.1999.5894. [DOI] [PubMed] [Google Scholar]
- 12.Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. Neuron. 1999;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- 13.Araneda RC, Kini AD, Firestein S. Nat Neurosci. 2000;3:1248–1255. doi: 10.1038/81774. [DOI] [PubMed] [Google Scholar]
- 14.de Bruyne M, Foster K, Carlson JR. Neuron. 2001;30:537–552. doi: 10.1016/s0896-6273(01)00289-6. [DOI] [PubMed] [Google Scholar]
- 15.Stensmyr MC, Giordano E, Balloi A, Angioy A-M, Hansson BS. J Exp Biol. 2003;206:715–724. doi: 10.1242/jeb.00143. [DOI] [PubMed] [Google Scholar]
- 16.Araneda RC, Peterlin Z, Zhang X, Chesler A, Firestein S. J Physiol. 2004;555:743–756. doi: 10.1113/jphysiol.2003.058040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shirokova E, Schmiedeberg K, Bedner P, Niessen H, Willecke K, Raguse J-D, Meyerhof W, Krautwurst D. J Biol Chem. 2005;280:11807–11815. doi: 10.1074/jbc.M411508200. [DOI] [PubMed] [Google Scholar]
- 18.Hallem EA, Carlson JR. Cell. 2006;125:143–160. doi: 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 19.Mori K, Takahashi YK, Igarashi KM, Yamaguchi M. Physiol Rev. 2006;86:409–433. doi: 10.1152/physrev.00021.2005. [DOI] [PubMed] [Google Scholar]
- 20.Schmuker M, de Bruyne M, Hähnel M, Schneider G. Chem Central J. 2007;1:11. doi: 10.1186/1752-153X-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korsching SI. Cell Mol Life Sci. 2001;58:520–530. doi: 10.1007/PL00000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korsching SI. Curr Opin Neurobiol. 2002;12:387–392. doi: 10.1016/s0959-4388(02)00348-3. [DOI] [PubMed] [Google Scholar]
- 23.Keller A, Vosshall LB. Curr Opin Neurobiol. 2003;13:103–110. doi: 10.1016/s0959-4388(03)00011-4. [DOI] [PubMed] [Google Scholar]
- 24.Friedrich RW, Korsching SI. Neuron. 1997;18:737–752. doi: 10.1016/s0896-6273(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 25.Uchida N, Takahashi YK, Tanifuji M, Mori K. Nat Neurosci. 2000;3:1035–1043. doi: 10.1038/79857. [DOI] [PubMed] [Google Scholar]
- 26.Meister M, Bonhoeffer T. J Neurosci. 2001;21:1351–1360. doi: 10.1523/JNEUROSCI.21-04-01351.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson BA, Farahbod H, Saber S, Leon M. J Comp Neurol. 2005;483:192–204. doi: 10.1002/cne.20415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuffenhauer A, Floersheim P, Acklin P, Jacoby E. J Chem Inf Comput Sci. 2003;43:391–405. doi: 10.1021/ci025569t. [DOI] [PubMed] [Google Scholar]
- 29.Kratochwil NA, Malherbe P, Lindemann L, Ebeling M, Hoener MC, Mühlemann A, Porter RHP, Stahl M, Gerber PR. J Chem Inf Model. 2005;45:1324–1336. doi: 10.1021/ci050221u. [DOI] [PubMed] [Google Scholar]
- 30.Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. Nat Biotechnol. 2007;25:197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 31.Cleland TA, Linster C. Chem Senses. 2005;30:801–813. doi: 10.1093/chemse/bji072. [DOI] [PubMed] [Google Scholar]
- 32.Linster C, Sachse S, Galizia CG. J Neurophysiol. 2005;93:3410–3417. doi: 10.1152/jn.01285.2004. [DOI] [PubMed] [Google Scholar]
- 33.Heisenberg M. Learn Mem. 1998;5:1–10. [PMC free article] [PubMed] [Google Scholar]
- 34.Zou Z, Horowitz LF, Montmayeur JP, Snapper S, Buck LB. Nature. 2001;414:173–179. doi: 10.1038/35102506. [DOI] [PubMed] [Google Scholar]
- 35.Marin EC, Jefferis GS X. E., Komiyama T, Zhu H, Luo L. Cell. 2002;109:243–255. doi: 10.1016/s0092-8674(02)00700-6. [DOI] [PubMed] [Google Scholar]
- 36.Wong AM, Wang JW, Axel R. Cell. 2002;109:229–241. doi: 10.1016/s0092-8674(02)00707-9. [DOI] [PubMed] [Google Scholar]
- 37.Roesch MR, Stalnaker TA, Schoenbaum G. Cereb Cortex. 2007;17:643–652. doi: 10.1093/cercor/bhk009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sigma–Aldrich. Flavors and Fragrances. Milwaukee, WI: Sigma–Aldrich; 2004. [Google Scholar]
- 39.Schmuker M, Schwarte F, Brück A, Proschak E, Tanrikulu Y, Givehchi A, Scheiffele K, Schneider G. J Mol Model. 2007;13:225–228. doi: 10.1007/s00894-006-0140-0. [DOI] [PubMed] [Google Scholar]
- 40.Kohonen T. Biological Cybernetics. 1982;V43:59–69. [Google Scholar]
- 41.Witten IH, Frank E. Data Mining: Practical Machine Learning Tools and Techniques. 2nd Ed. San Francisco: Morgan Kaufmann; 2005. [Google Scholar]
- 42.Laurent G. Nat Rev Neurosci. 2002;3:884–895. doi: 10.1038/nrn964. [DOI] [PubMed] [Google Scholar]
- 43.Mjolsness E, DeCoste D. Science. 2001;293:2051–2055. doi: 10.1126/science.293.5537.2051. [DOI] [PubMed] [Google Scholar]
- 44.Schneider P, Schneider G. QSAR Comb Sci. 2003;22:713–718. [Google Scholar]
- 45.Latino DA, Aires-de-Sousa J. Angew Chem Int Ed Engl. 2006;45:2066–2069. doi: 10.1002/anie.200503833. [DOI] [PubMed] [Google Scholar]
- 46.Huerta R, Nowotny T, Garciá-Sanchez M, Abarbanel HDI, Rabinovich MI. Neural Comput. 2004;16:1601–1640. doi: 10.1162/089976604774201613. [DOI] [PubMed] [Google Scholar]
- 47.Nowotny T, Huerta R, Abarbanel HDI, Rabinovich MI. Biol Cybern. 2005;93:436–446. doi: 10.1007/s00422-005-0019-7. [DOI] [PubMed] [Google Scholar]
- 48.Meissner M, Schmuker M, Schneider G. BMC Bioinformatics. 2006;7:125. doi: 10.1186/1471-2105-7-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Halgren TA. J Comp Chem. 1999;20:720–729. doi: 10.1002/(SICI)1096-987X(199905)20:7<720::AID-JCC7>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]