Abstract
Accurate determination of the relative positions of proteins within localized regions of the cell is essential for understanding their biological function. Although fluorescent fusion proteins are targeted with molecular precision, the position of these genetically expressed reporters is usually known only to the resolution of conventional optics (≈200 nm). Here, we report the use of two-color photoactivated localization microscopy (PALM) to determine the ultrastructural relationship between different proteins fused to spectrally distinct photoactivatable fluorescent proteins (PA-FPs). The nonperturbative incorporation of these endogenous tags facilitates an imaging resolution in whole, fixed cells of ≈20–30 nm at acquisition times of 5–30 min. We apply the technique to image different pairs of proteins assembled in adhesion complexes, the central attachment points between the cytoskeleton and the substrate in migrating cells. For several pairs, we find that proteins that seem colocalized when viewed by conventional optics are resolved as distinct interlocking nano-aggregates when imaged via PALM. The simplicity, minimal invasiveness, resolution, and speed of the technique all suggest its potential to directly visualize molecular interactions within cellular structures at the nanometer scale.
Keywords: fluorescent proteins, multi-label
The dynamic functionality of the cell is driven by interactions between its molecular constituents. A key advantage of fluorescence microscopy is its ability to map the relative distribution of two or more of these constituents by labeling them with spectrally distinct labels (1). However, many of the interactions occur in localized regions too small to be accurately probed with the limited resolution of conventional optics (≈200 nm).
In response, several superresolution multicolor fluorescence methods have been developed. Near-field microscopy, for example, has been used to map the relative distribution of lipid patches and membrane proteins in fibroblasts (2), as well as the colocalization of malarial and host skeletal proteins in erythrocytes (3). Stimulated emission depletion microscopy has been used to image synaptic and mitochondrial proteins (4). The fluorescent tags FlAsH and ReAsH can be viewed at diffraction-limited resolution, after which ReAsH can initiate a photo-oxidation reaction for subsequent correlative electron microscopic imaging at much higher resolution (5).
Near-molecular resolution can be achieved by another means of superresolution that relies on the optical isolation of individual molecules within a densely labeled specimen, followed by the statistical estimation of their respective coordinates (6). Initially demonstrated (7, 8) by using genetically expressed photoactivatable fluorescent proteins (PA-FPs) (9) at the high molecular density (>105/μm2) and localization precision required for macromolecular resolution, the same concept has subsequently been applied, although at lower density, using various fluorescent reporters (10–13). Recently, two methods of multicolor imaging based on this concept have been described: the first using photoswitching in the FP rsFastLime and the organic fluorophore Cy5 (both targeted to β-tubulin) (14), and the second using Cy dyes in activator/reporter pairs targeted to microtubules and clathrin-coated pits (15). In both cases, the photoswitchable labels were exogenously introduced via antibodies (Abs) or biotinylation.
Here, however, we report two-color photoactivated localization microscopy (PALM) using the endogenously expressed PA-FP pairs Dronpa (16)/EosFP (17) and PS-CFP2 (18)/EosFP. This approach offers several key advantages. First, PA-FPs can be viewed in living cells by conventional epi-fluorescence techniques and then fixed for PALM imaging at a physiologically significant time. Second, PA-FPs should be compatible with eventual live-cell PALM imaging. Third, sample preparation consists of only mild fixation, after which the cells are returned to physiological media; no potentially perturbative detergents, oxygen scavenging agents, or treatments to manipulate the molecular photophysics need be applied. Fourth, PA-FPs are expressed bound to their respective targets, and therefore the specificity and background issues that must be addressed with exogenous labeling are eliminated. Fifth, this binding occurs on a scale (≈1–2 nm) consistent with the localization precision possible in PALM. Abs, however, add considerable uncertainty (≈10–20 nm) in the spatial relationship between the label and its target (particularly with secondary Abs), and thus a small molecule labeling strategy (19) will be needed to achieve optimal resolution when using exogenous fluorophores. Finally, although it is necessary to guard against overexpression of the target protein and the effect of the PA-FP tag on function and organization of this protein, higher labeling densities can in principle be achieved with PA-FPs than with Abs [supporting information (SI) Fig. 6], because of the significantly smaller size of the former (≈2 nm vs. ≈10 nm). Molecular proximity is an important consideration for PALM, where the density of localized molecules is as significant as the precision of their localization in determining the ultimate spatial resolution. To illustrate these advantages, we demonstrate two-color PALM with endogenous PA-FP labels by applying it to study the ultrastructural relationship between different pairs of proteins involved in adhesion complexes (20), transmembrane cytoskeleton-substrate attachment points central to cell migration.
Results and Discussion
Determination of Suitable PA-FP Pairs.
Several constraints guide the selection of PA-FP pairs suitable for dual-color PALM. Most obvious is the need for spectrally distinct labels. However, whereas the palette of conventional FPs spans much of the visible spectrum (21), most current PA-FPs (9, 22) fall into two classes: those that emit green fluorescence when photoactivated (e.g., PA-GFP, PS-CFP, and Dronpa) and those whose emission shifts from green-to-orange (e.g., KikGR (23), and EosFP). Using one of each class as a dual-label pair is problematic, because the large pool of preactivated green molecules from the green-to-orange species forms a bright background that impedes the isolation and localization of single photoactivated green molecules of the other type.
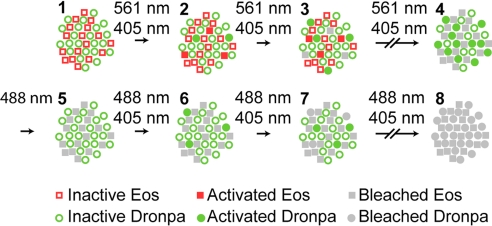
One solution introduced here relies on the reversible photoswitching characteristics of Dronpa. Thus, as conceptualized in Fig. 1, Dronpa is coexpressed with a green-to-orange PA-FP, usually EosFP in the tandem dimer form (tdEos). Both species are initially in their inactive state (Fig. 1, step 1). The tdEos molecules are then serially isolated, localized, and bleached over the course of thousands of image frames (steps 2 and 3) by means of continuous application of activation and excitation light (405 and 561 nm, respectively). Eventually, the pool of tdEos molecules is exhausted (step 4). However, the prolonged exposure to the activation light leaves a large pool of activated Dronpa molecules. To reduce the intense green background associated with these molecules, 488 nm light is applied to deactivate most of them (step 5), until single Dronpa molecules can be discerned. Activation and excitation light (405 and 488 nm, respectively) are then reapplied (steps 6 and 7) until all remaining Dronpa molecules are isolated, localized, and bleached (step 8). Finally, the aggregate localization data from tdEos and Dronpa are mutually aligned using similar data measured from luminescent Au fiducial beads (8) that are indefinitely photostable and visible in both detection channels. The result is a dual-color PALM image of the relative distribution of the two target proteins to which the tdEos and Dronpa are bound.
Fig. 1.
Protocol for dual-label superresolution imaging by PALM. A specimen initially expressing inactive EosFP and Dronpa molecules (step 1) is exposed to a 405-nm activation light and a 561-nm Eos-excitation light (steps 2 and 3) until all EosFP molecules are detected, localized, and bleached (step 4). The many active Dronpa molecules that then exist (step 4) are deactivated by using an intense 488-nm light (step 5). Both 405-nm and 488-nm light are then applied (steps 6 and 7) to serially activate, detect, localize, and eventually bleach (step 8) all remaining Dronpa molecules. PALM images encompassing 105 to 106 molecules are thereby acquired, typically in 5–30 min each.
Another characteristic critical to the success of PALM is the contrast ratio between the activated and inactivated forms of the photoswitchable label at the excitation wavelength of the activated form. Within the more densely packed areas of the specimen, thousands of labeled molecules can reside in a single diffraction limited region (DLR). If the contrast ratio is not at least as large, the individually weak emission from the many inactive molecules in the DLR can together overwhelm the emission from a single activated molecule, rendering the localization of the latter inaccurate or even impossible. This effect is illustrated in SI Fig. 7, where PALM images taken with PA-FPs of differing contrast ratios are compared. One could limit the application of the technique to less densely labeled regions or prebleach the specimen until the labeling density matches the contrast ratio. However, these solutions result in significantly lower spatial resolution, as the Nyquist-Shannon theorem (24) requires the sampling interval (i.e., the mean distance between neighboring localized molecules) to be at least twice as fine as the desired resolution. EosFP has the highest contrast ratio of all of the PA-FPs we have tested (SI Fig. 7A) and has been shown to be suitable for PALM imaging at densities of >105 molecules per μm2 (8). Therefore, it was chosen here as the green-to-orange PA-FP to pair with Dronpa for dual-label PALM.
Minimal Crosstalk Between PA-FP Pairs.
A key metric by which to judge PA-FP pairs for PALM is the extent to which each label correctly identifies its designated target species. This criterion is assessed by means of dual-channel crosstalk experiments in SI Fig. 8. Thus, in SI Fig 8A, a human foreskin fibroblast (HFF-1) cell transfected with tdEos-paxillin only was first PALM-imaged for 100,000 frames under activation and excitation (561 nm) appropriate for EosFP (SI Fig. 8A Left) until few molecules remained. The same cell was then PALM-imaged for 50,000 frames under activation and excitation (488 nm) appropriate for Dronpa, even though no Dronpa existed (SI Fig 8A Right). Comparison of the figures, plotted at identical gain, reveals negligible misidentification of the EosFP as Dronpa.
The converse experiment is shown in SI Fig. 8B: an HFF-1 cell transfected with Dronpa-paxillin only was first PALM-imaged for 50,000 frames under conditions appropriate for EosFP (Left), followed by PALM-imaging under conditions suitable for Dronpa (Right). As expected, adhesion structures are visible only in the Dronpa channel. Indeed, the number of molecules in the Eos channel is comparable with that seen in control experiments on untransfected cells.
Performance of Dual-Label PALM.
To illustrate the utility of dual label PALM in a biologically relevant context, adhesion complexes (ACs) in HFF-1 cells were studied. These intricate structures can be as small as 0.5 μm and result from the nanoscale interaction of >90 different proteins (25). Therefore, multilabel PALM has the potential to play an important role in deciphering their organization and the process of their formation. Conversely, ACs are well suited to PALM study because they contain proteins densely concentrated in the intra- and extracellular space near the cell–substrate interface (20). Thus, not only is the molecular density consistent with PALM resolution on the macromolecular (≈10–20 nm) level, but many of the constituents are within the evanescent field of total internal reflection fluorescence (TIRF) excitation, and thus can be viewed without significant background autofluorescence or the confusion that results when a deeply three-dimensional structure is projected into a two-dimensional image space.
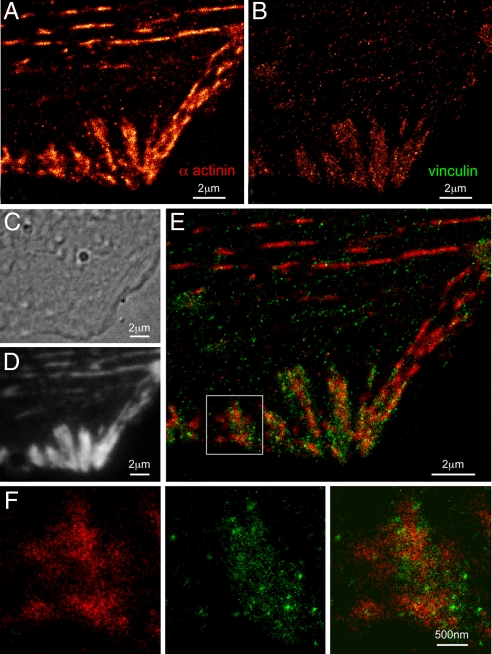
An example, showing the nanoscale organization of Dronpa-tagged α-actinin and tdEos-tagged vinculin in the same HFF-1 cell, is given in Fig. 2. α-Actinin (Fig. 2 A and F) is a protein that cross-links actin filaments within stress fibers and ventral actin bundles of motile cells and associates with numerous proteins involved in ACs, including vinculin (26). The large apparent variation in α-actinin density along many stress fibers in Fig. 2A may reflect variations in the distances of such fibers from the substrate and therefore their locations within the evanescent excitation field. Vinculin (Fig. 2 B and F), although not essential for FA assembly, can bind α-actinin and AC proteins such as paxillin, talin, and VASP. Indeed, it is believed to stabilize interactions between talin and actin, and is known to localize at the termini of stress fibers (27). This feature is apparent in both the dual-label PALM overlay (Fig. 2 E and F) and diffraction-limited TIRF (Fig. 2D) images of the cell. However, only the PALM data reveals that α-actinin and vinculin do not completely colocalize within each AC, a result in agreement with the partial correlation of their retrograde movement observed in refs. 28 and 29. Instead, α-actinin exists in larger patches emanating from the stress fibers, whereas vinculin coalesces in small, dense clusters scattered throughout each adhesion.
Fig. 2.
Nanostructural organization of cytoskeletal α-actinin and adhesion complex localized vinculin in an HFF-1 cell. (A) PALM image of Dronpa-tagged α-actinin. (B) PALM image of tdEos-tagged vinculin. (C) DIC image revealing morphology. (D) TIRF image of combined tdEos and Dronpa emission. (E) Dual-color PALM overlay of α-actinin (red) and vinculin (green). (F) α-Actinin, vinculin, and overlaid PALM images within the single adhesion shown in the box in E. E and F reveal that α-actinin and vinculin only partially colocalize within each adhesion, with α-actinin existing in large patches emanating from stress fibers and vinculin coalescing in small, dense clusters scattered across each adhesion.
Fig. 2A and Fig. 2B are sufficiently distinct to underscore the low crosstalk in opposite detection channels for EosFP and Dronpa. Fig. 2 A and B also indicates that a similar number of molecules can be localized (105 to 106 per PALM image) with each label, even though Dronpa is imaged after prolonged exposure to 405 nm and 561 nm light, followed by deactivation with 488 nm light. Finally, Fig. 2 C and D demonstrates that the sample preparation protocol does not perturb the cellular morphology and membrane structure, at least at the diffraction-limited level of differential interference contrast (DIC) and TIRF.
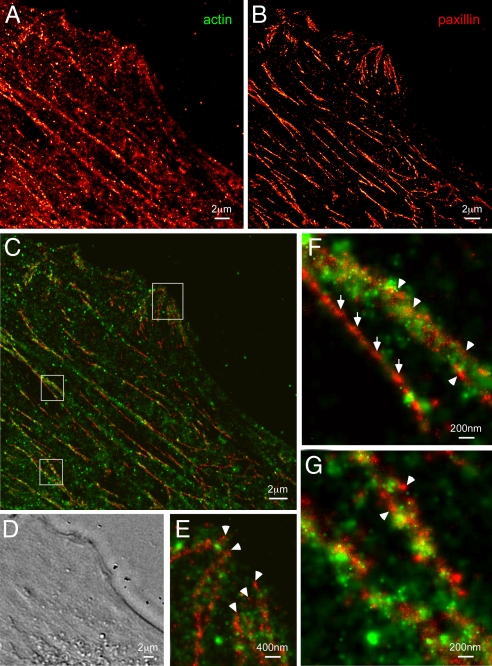
Another example illustrating the nanoscale relationship between a cytoskeletal protein (Dronpa-tagged actin) and an adhesion-associated protein (tdEos-tagged paxillin) is given in Fig. 3. The Dronpa image (Fig. 3A) reveals not only ventral actin fibers that span the length of the HFF-1 cell, but also a finer actin network amid these fibers that is typically not visible by TIRF. Paxillin (Fig. 3B) exists not only in ACs at the cell periphery (e.g., Fig. 3E), but also as elongated adhesive complexes of order 100 nm in diameter or less running parallel to the ventral actin fibers (Fig. 3C).
Fig. 3.
Nanostructural organization of cytoskeletal actin and the adhesion protein paxillin in an HFF-1 cell. (A) PALM image of Dronpa-tagged actin. (B) PALM image of tdEos-tagged paxillin. (C) Dual-color PALM overlay of actin (green) and paxillin (red). (D) DIC image in the same region. Paxillin assembles in fibrillar-like adhesions that run parallel to ventral actin fibers and cluster near the cell periphery to form larger adhesion complexes (topmost box in C, shown expanded in E). Very little direct overlap is observed between actin and paxillin, although actin bundles densely cluster around some (arrowheads in E–G) but not all (full arrows in F) fibrillar paxillin adhesions.
In contrast to the vinculin/α-actinin example in Fig. 2, there is very little direct overlap between paxillin and actin (Fig. 3 C and E–G). This segregation is consistent with the relatively low correlation observed in the retrograde movement of these two proteins within adhesion complexes (28, 29). Nevertheless, higher magnification views indicate that some (arrow heads, Fig. 3 E–G), but not all (full arrows, Fig. 3F) of the long paxillin structures form an inner core surrounded by bundled actin fibers. This structural relationship would not be discernable by two-color TIRF.
One obvious qualitative difference between the Dronpa and tdEos images in Fig. 3 A and B, respectively, is that the latter exhibits substantially higher resolution. This difference is quantified in SI Fig. 9A, where histograms are given of the precision to which the molecules in these images are localized (30). EosFP is localized on average more than twice as precisely as Dronpa. The greater localization precision reflects both the higher active/inactive contrast ratio of EosFP, which results in lower background (SI Fig. 9B), and the inadvertent deactivation of Dronpa, which leads to fewer collected photons per molecule per activation event (SI Fig. 9C).
PS-CFP2 as an Alternative Second Label.
When imaging EosFP and Dronpa by the protocol in Fig. 1, after completion of the Eos imaging (step 4), the pool of activated Dronpa molecules and associated unwanted fluorescence background was far lower than expected. Although several explanations for this phenomenon remain to be explored (e.g., light-induced or spontaneous deactivation), it raised the question as to whether other, ostensibly nonreversible, green PA-FPs might prove suitable for double-label PALM. In particular, PS-CFP2 is reported (18) to have an on/off contrast ratio nearly as good as EosFP, and therefore was chosen to explore this possibility.
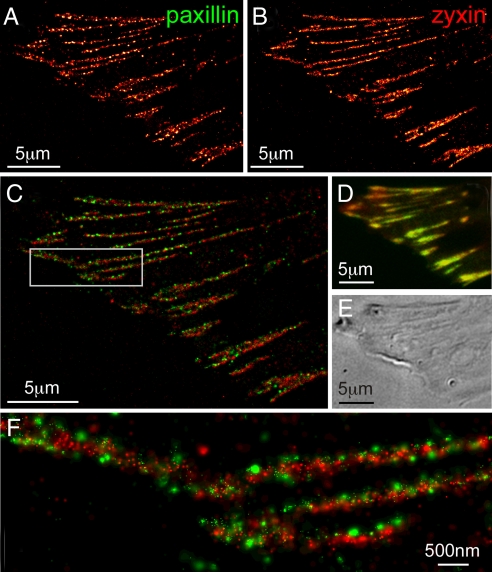
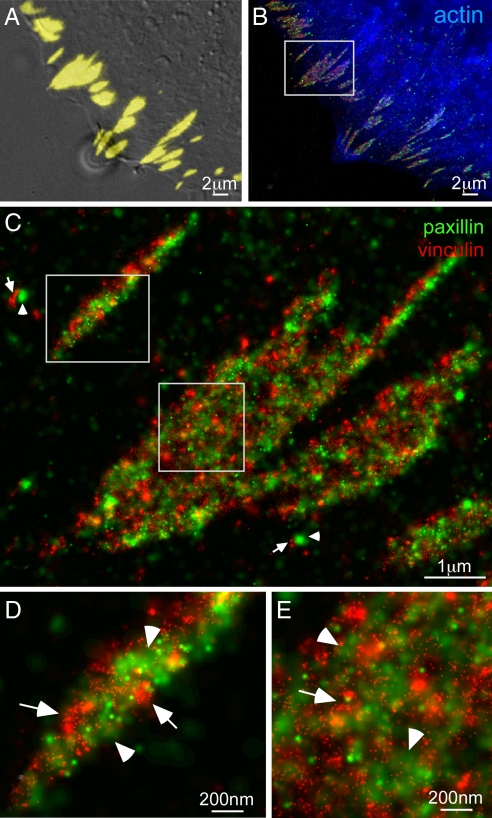
The results, shown in Fig. 4, show that this approach is indeed viable: tdEos-tagged paxillin (Fig. 4A) and PS-CFP2-tagged zyxin (Fig. 4B), which seem colocalized (31) when viewed by conventional TIRF (Fig. 4D) are clearly resolved as separate nanoscale clusters that assemble across each AC (Fig. 4 C and F). It is true that the background from activated PS-CFP2 immediately after imaging Eos (step 4 in Fig. 1) is consistently much brighter than when Dronpa is used. Nevertheless, in nearly all attempts this background could be bleached and/or deactivated (32) by exposure to 488 nm excitation, after which the remaining pool of unconverted PS-CFP2 molecules was still sufficiently large to generate PALM images at acceptable molecular densities.
Fig. 4.
Dual-color PALM using PS-CFP2 as an alternative second label. (A) PALM image of tdEos-tagged paxillin. (B) PALM image of PS-CFP2-tagged zyxin. (C) Dual-color PALM overlay of paxillin (green) and zyxin (red). (D) Diffraction-limited, summed molecule, dual-color TIRF image (8). (E) DIC image. The two adhesion proteins seem colocalized in D, but are revealed in C and the boxed region shown at higher magnification in F to have very little overlap, with paxillin tightly clustered in separate nano-domains.
Furthermore, comparison of Figs. 3A and 4B qualitatively suggests that higher spatial resolution can be obtained with PS-CFP2 than with Dronpa. However, this suggestion is at odds with SI Fig. 9A, where similar localization precision is calculated for the two labels: on average the background for Dronpa is less than half as large (SI Fig. 9B), but PS-CFP2 emits nearly 5-fold as many photons per molecule (SI Fig. 9C). The implication is that the true localization precision for Dronpa is worse than calculations would suggest. This conclusion may reflect the fact that, when calculating the precision, the background noise is assumed to be Poisson distributed (30), whereas for Dronpa additional noise may arise from spontaneous photoswitching between its two states.
Triple-Label Imaging with mCerulean.
Dual-label PALM can also be used in conjunction with conventional FPs spectrally distinct from Eos and Dronpa/PS-CFP2 to yield hybrid images of multiple proteins combining both diffraction-limited and superresolution information. For example, in Fig. 5B, PALM images of Dronpa-tagged paxillin and tdEos-tagged vinculin overlaid with an epi-fluorescence image of mCerulean-tagged actin in the same HFF-1 cell shows adhesion complexes aligned with and at the termini of actin bundles. Although previous (33) diffraction-limited imaging has shown that paxillin and vinculin colocalize in ACs (as in Fig. 5A), dual-color PALM reveals (Fig. 5 C–E) parallel arrays of interwoven but noncolocalized paxillin and vinculin aggregates along the length of each AC. Furthermore, even when the Au bead-based alignment used here is abandoned in an artificial attempt to maximize overlap by cross-correlation or manual means, the degree of colocalization between the proteins remains poor.
Fig. 5.
Triple-label imaging with combined techniques. (A) Overlaid DIC and TIRF (yellow) images of paxillin and vinculin coexpressed in an HFF-1 cell. (B) Diffraction-limited epi-fluorescence image of mCerulean-tagged actin (blue) overlaid with PALM images of Dronpa-tagged paxillin (green) and tdEos-tagged vinculin (red) shows adhesion complexes at the periphery of the cell aligned with the termini of actin bundles. An expanded view (C) of the boxed region in B reveals parallel arrays of interwoven paxillin and vinculin aggregates along the length of each AC, as well as possibly nascent adhesion complexes consisting of adjacent paxillin (arrowheads) and vinculin aggregates (arrows). Further magnified views (D and E) of the boxed regions in C indicate other examples of adjacent aggregates of either paxillin (arrowheads) or vinculin (arrows) within larger adhesions.
Summary.
The above results clearly show that two-color PALM can reveal the spatial relationship between two proteins in whole, fixed cells at all length scales down to the nanometric level (≈20–30 nm). Furthermore, such results are obtained routinely in 5–30 min on a live-cell compatible commercial microscope equipped with DIC, TIRF, and epi-fluorescence optics, permitting cell morphologies, transfection levels, and spatial distributions of conventional FP-tagged proteins to be measured as well.
Nevertheless, considerable room remains for improvement. First, neither Dronpa nor PS-CFP2 can be localized as precisely as EosFP. Second, the approach requires the two labels to be imaged serially, so it is unsuited to live-cell imaging. Finally, there remains the risk that Dronpa or PS-CFP2 molecules may be bleached while imaging EosFP, eliminating valuable data. For all these reasons, we continue to search for new, spectrally distinct PA-FPs with high on/off contrast ratios to pair with existing PA-FPs for multilabel PALM.
A striking feature present in many of the PALM images is the apparent aggregation of the molecules in numerous clusters scattered throughout the adhesion complexes. Such aggregates may be natural and exist throughout the entire volume of the ACs, or they may represent points where continuous protein filaments dive in and out of the evanescent excitation field. Certainly, some of the periodic light/dark patterns seen in stress fibers (Fig. 2A) and fibrillar-like ACs (Fig. 4 A and B) suggest this latter possibility. Interestingly, recent atomic force microscopy images of “de-roofed” fibroblasts cells also reveal adhesions consisting of many fibrillar-like structures decorated with numerous globular aggregates, all of dimensions (20–80 nm) similar to those seen here (34). However, a more disquieting option is that aggregation occurs because of the PA-FP tags, and that untagged proteins would not exhibit this behavior.
Despite this caveat, cells imaged 24–48 h after transfection exhibited very similar motility, growth, and morphology to their untransfected cousins and, at the diffraction limit, PA-FP-expressing cells appeared similar to those expressing conventional FP tags. It is therefore tempting to draw some tentative conclusions concerning AC organization from our results. First, different AC proteins that were previously thought to completely overlap based on quantitative diffraction limited microscopy are revealed by PALM to organize into nano-aggregates that in turn assemble into higher order structures. Second, proteins that belong to similar functional groups (e.g., vinculin and α-actinin in Fig. 2), and have some degree of coordinated retrograde movement within ACs (28, 29), exhibit a certain amount of nanostructural overlap. In contrast, proteins that are from functionally distinct groups (e.g., paxillin and actin in Fig. 3), and have almost no coherent movement, show very little overlap. Finally, and perhaps most significantly, within adhesive structures there is a remarkable amount of interdigitation of different protein nano-aggregates, creating an interwoven arrangement. These findings suggest a much more complex and interactive organization than previously inferred by conventional means, and indicate the potential of multicolor PALM to address a multitude of questions at the interface between molecular and cellular biology.
Materials and Methods
Instrumentation.
PALM imaging was performed on an Olympus IX81 inverted microscope equipped with DIC optics and an internal 200-mm focal length relay lens to focus laser light at the rear pupil of a ×100, 1.65 N.A. objective. Laser light was delivered to the microscope through free space from a platform where 405-nm, 488-nm, and 561-nm lasers were combined (SI Fig. 10). Single-molecule Dronpa, PS-CFP2, or Eos fluorescence signals generated during acquisition were separated from the activation and excitation light using appropriate filter sets within the microscope (SI Table 1) and passed to an electron-multiplying CCD camera for detection. Sample drift in each channel was corrected by tracking the motion of 40- and 100-nm-diameter Au fiducial beads (790114-010 and 790122-010; Microspheres-Nanospheres) added to the sample before PALM imaging (8). Images from the two channels were aligned by recording the position of a fiducial bead common to both channels. Localization and image-rendering algorithms were as described elsewhere (8). Using continuously applied activation and typical excitation intensities of ≈0.5–2.0 kW/cm2, single-molecule frame times of 20 to 50 ms were common, yielding complete PALM images every 5–30 min. Further information is given in SI Materials and Methods.
Sample Preparation.
HFF-1 cells (ATCC, SCRC-1041) passages 17–26 were grown in DMEM-HG containing 15% FBS to 55–85% confluency. Trypsinized cells were then transiently transfected at ≈2–4 × 105 cells per well with a Nucleofector 96-well shuttle system (Amaxa Biosystems) using Cell Line Nucleofector Kit SF, program DS-137, and ≈0.3–1.0 μg per well plasmid DNA. Plasmid construction is detailed in SI Materials and Methods. Separately, high refractive index coverslips (Olympus, APO100X-CG) were (i) incubated in 5:1:1 Milli-Q filtered H2O:ammonium hydroxide:hydrogen peroxide for ≈12 h at 75°C; (ii) serially rinsed in H2O and methanol; (iii) flamed; (iv) coated overnight with 5–10 μg/ml fibronectin at 4°C; and (v) blocked with heat-inactivated 1% BSA for 1 h at 37°C. Transfected cells were then (i) transferred to the coverslips; (ii) grown as above for 24–36 h; (iii) fixed for 15 min at 37°C in 2% paraformaldehyde in PHEM (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, and 2 mM MgCl2, pH 6.9); and (iv) rinsed 3× with PHEM. To compensate for sample drift during acquisition, cells were then incubated 15–30 min with 40 and 100 nm Au beads in suspension diluted 10× in PHEM, before a final rinse with PHEM.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Nima Ghitani for assistance with fixation and antibody staining; Kevin McGowan, Christopher Murphy, Anna Ozarowska, and Patrice Worthy for help with plasmid construction, characterization, and amplification; George Patterson (NIH/NICHD, Bethesda, MD) for the gift of caged-fluorescein secondary antibodies; Na Ji for critically reading the manuscript; and Clare Waterman-Storer for advice and encouragement regarding the adhesion complex system. We also thank Jörg Wiedenmann (University of Ulm, Ulm, Germany), Tom Keller (Florida State University, Tallahassee, FL), Rick Horwitz (University of Virginia, Charlottesville), Clare Waterman-Storer (National Institutes of Health/National Heart, Lung, and Blood Institute), David Piston (Vanderbilt University, Nashville, TN), and George Patterson for the gift of various plasmids. This research was supported in part by the Intramural Research Programs of the National Institutes of Health, the National Institute of Dental and Craniofacial Research, the National Institute of Neurological Disorders and Stroke, and the National Institute of Child Health and Human Development.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710517105/DC1.
References
- 1.Giepmans BNG, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 2.Hwang J, Gheber LA, Margolis L, Edidin M. Domains in cell plasma membranes investigated by near-field scanning optical microscopy. Biophys J. 1998;74:2184–2190. doi: 10.1016/S0006-3495(98)77927-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Enderle T, Ha T, Ogletree DF, Chemla DS, Magowan C, Weiss S. Membrane specific mapping and colocalization of malarial and host skeletal proteins in the Plasmodium falciparum infected erythrocyte by dual-color near-field scanning optical microscopy. Proc Natl Acad Sci USA. 1997;94:520–525. doi: 10.1073/pnas.94.2.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donnert G, Keller J, Wurm CA, Rizzoli SO, Westphal V, Schönle A, Jahn R, Jakobs S, Eggeling C, Hell SW. Two-color far-field fluorescence nanoscopy. Biophys J. 2007;92:L67–L69. doi: 10.1529/biophysj.107.104497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaietta G, Deerinck TJ, Adams SR, Bouwer J, Tour O, Laird DW, Sosinsky GE, Tsien RY, Ellisman MH. Multicolor and electron microscopic imaging of connexin trafficking. Science. 2002;296:503–507. doi: 10.1126/science.1068793. [DOI] [PubMed] [Google Scholar]
- 6.Betzig E. Proposed method for molecular optical imaging. Opt Lett. 1995;20:237–239. doi: 10.1364/ol.20.000237. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y-l, Hahn KM, Murphy RF, Horwitz AF. From imaging to understanding: Frontiers in live cell imaging. J Cell Biol. 2006;174:481–484. doi: 10.1083/jcb.200607097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 9.Wiedenmann J, Nienhaus GU. Live-cell imaging with EosFP and other photoactivatable marker proteins of the GFP family. Exp Rev Proteom. 2006;3:361–374. doi: 10.1586/14789450.3.3.361. [DOI] [PubMed] [Google Scholar]
- 10.Hess ST, Girirajan TPK, Mason MD. Ultrahigh resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Methods. 2006;3:793–796. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharonov A, Hochstrasser RM. Wide-field subdiffraction imaging by accumulated binding of diffusing probes. Proc Natl Acad Sci USA. 2006;103:18911–18916. doi: 10.1073/pnas.0609643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egner A, Geisler C, von Middendorff C, Bock H, Wenzel D, Medda R, Andresen M, Stiel AC, Jakobs S, Eggeling C, et al. Fluorescence nanoscopy in whole cells by asynchronous localization of photoswitching emitters. Biophys J. 2007;93:3285–3290. doi: 10.1529/biophysj.107.112201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bock H, Geisler C, Wurm CA, von Middendorff C, Jakobs S, Schönle A, Egner A, Hell SW, Eggeling C. Two-color far-field fluorescence nanoscopy based on photoswitchable emitters. Appl Phys B. 2007;88:161–165. [Google Scholar]
- 15.Bates M, Huang B, Dempsey GT, Zhuang X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science. 2007;317:1749–1753. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ando R, Mizuno H, Miyawaki A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 2004;306:1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- 17.Wiedenmann J, Ivanchenko S, Oswald F, Schmitt F, Röcker C, Salih A, Spindler KD, Nienhaus GU. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc Natl Acad Sci USA. 2004;101:15905–15910. doi: 10.1073/pnas.0403668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chudakov DM, Verkhusha VV, Staroverov DB, Souslova EA, Lukyanov S, Lukyanov KA. Photoswitchable cyan fluorescent protein for protein tracking. Nat Biotechnol. 2004;22:1435–1439. doi: 10.1038/nbt1025. [DOI] [PubMed] [Google Scholar]
- 19.Chen I, Ting AY. Site-specific labeling of proteins with small molecules in live cells. Curr Opin Biotechnol. 2005;16:35–40. doi: 10.1016/j.copbio.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Zamir E, Geiger B. Molecular complexity and dynamics of cell-matrix adhesions. J Cell Sci. 2001;114:3583–3590. doi: 10.1242/jcs.114.20.3583. [DOI] [PubMed] [Google Scholar]
- 21.Shaner N, Steinbach P, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 22.Lukyanov KA, Chudakov DM, Lukyanov S, Verkhusha VV. Photoactivatable fluorescent proteins. Nat Rev Mol Cell Biol. 2005;6:885–890. doi: 10.1038/nrm1741. [DOI] [PubMed] [Google Scholar]
- 23.Tsutsui H, Karasawa S, Shimizu H, Nukina N, Miywaki A. Semirational engineering of a coral fluorescent protein into an efficient highlighter. EMBO Rep. 2005;6:233–238. doi: 10.1038/sj.embor.7400361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shannon CE. Communication in the presence of noise. Proc Inst Radio Eng. 1949;37:10–21. [Google Scholar]
- 25.Zaidel-Bar R, Itzkovitz S, Ma'ayan A, Iyengar R, Geiger B. Functional atlas of the integrin adhesome. Nat Cell Biol. 2007;9:858–867. doi: 10.1038/ncb0807-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otey CA, Carpen O. Alpha-actinin revisited: A fresh look at an old player. Cell Motil Cytoskel. 2004;58:104–111. doi: 10.1002/cm.20007. [DOI] [PubMed] [Google Scholar]
- 27.Ziegler WH, Liddington RC, Critchley DR. The structure and regulation of vinculin. Trends Cell Biol. 2006;16:453–460. doi: 10.1016/j.tcb.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Hu K, Ji L, Applegate KT, Danuser G, Waterman-Storer CM. Differential transmission of actin motion within focal adhesions. Science. 2007;315:111–115. doi: 10.1126/science.1135085. [DOI] [PubMed] [Google Scholar]
- 29.Brown CM, Hebert B, Kolin DL, Zareno J, Whitmore L, Horwitz AR, Wiseman PW. Probing the integrin-actin linkage using high-resolution protein velocity mapping. J Cell Sci. 2006;119:5204–5214. doi: 10.1242/jcs.03321. [DOI] [PubMed] [Google Scholar]
- 30.Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaidel-Bar R, Ballestrem C, Kam Z, Geiger B. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J Cell Sci. 2003;116:4605–4613. doi: 10.1242/jcs.00792. [DOI] [PubMed] [Google Scholar]
- 32.Chudakov DM, Lukyanov S, Lukyanov KA. Tracking intracellular protein movements using photoswitchable fluorescent proteins PS-CFP2 and Dendra2. Nat Protoc. 2007;2:2024–2032. doi: 10.1038/nprot.2007.291. [DOI] [PubMed] [Google Scholar]
- 33.Zamir E, Katz BZ, Aota S, Yamada KM, Geiger B, Kam Z. Molecular diversity of cell-matrix adhesions. J Cell Sci. 1999;112:1655–1669. doi: 10.1242/jcs.112.11.1655. [DOI] [PubMed] [Google Scholar]
- 34.Franz CM, Muller DJ. Analyzing focal adhesion structure by atomic force microscopy. J Cell Sci. 2005;118:5315–5323. doi: 10.1242/jcs.02653. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.