Abstract
Soj is a member of the ParA family of ATPases involved in plasmid and chromosomal segregation. It binds nonspecifically and cooperatively to DNA although the function of this binding is unknown. Here, we show that mutation of conserved arginine residues that map to the surface of Bacillus subtilis Soj caused only minimal effects on nucleotide-dependent dimerization but had dramatic effects on DNA binding. Using a model plasmid partitioning system in Escherichia coli, we find that Soj DNA-binding mutants are deficient in plasmid segregation. The location of the arginines on the Soj structure explains why DNA binding depends on dimerization and was used to orient the Soj dimer on the DNA, revealing the axis of Soj polymerization. The arginine residues are conserved among other chromosomal homologues, including the ParAs from Caulobacter crescentus, Pseudomonas aeruginosa, Pseudomonas putida, Streptomyces coelicolor, and chromosome I of Vibrio cholerae indicating that DNA binding is a common feature of members of this family.
The stable maintenance of low-copy plasmids in bacteria is achieved through the action of partitioning proteins encoded within par (for partitioning) loci (1, 2). These loci are composed of two genes, parA/F/M and parB/G/R, and one or more cis-acting sites, parS or parC. parA/F/M encodes an ATPase (ParA/F/M) and parB/G/R encodes a DNA-binding protein (ParB/G/R), which specifically binds to parS or parC. All three components are required for maintenance of low-copy plasmids. Bacterial chromosomes also have par loci, although they are notably absent from the genome of Escherichia coli and its close relatives. The chromosomal parAB locus of Caulobacter crescentus is essential for growth (3), whereas in Bacillus subtilis, Pseudomonas putida, Pseudomonas aeruginosa, Streptomyces coelicolor, and Vibrio cholerae (chromosome I parAB), the locus is not essential. In these organisms, disruption primarily causes segregation defects during developmental shifts such as sporulation or entry into stationary phase (4–8).
Based on the sequence of the ATPase, par loci have been divided into two groups types I and II (9, 10). In type I, the ATPase, called ParA, Soj, or ParF, is related to MinD, a protein involved in spatial regulation of cell division. It includes chromosomal and plasmid-encoded loci. The plasmid-encoded loci are subdivided further: Type Ia have a larger ParA because of an N-terminal extension involved in specific DNA binding; type Ib have a smaller ParA that lacks the N-terminal extension. Chromosomal ParAs are similar to type Ib. In type II loci, the ATPase, called ParM, is related to actin, and the understanding of how these loci function to mediate plasmid segregation is more advanced. Studies in vivo demonstrate that ParM from R1 plasmid forms filaments that can extend the length of the cell and have plasmids associated with their ends (11, 12). Studies in vitro reveal that ParM filaments undergo ATP-dependent dynamic instability but are stabilized in the presence of plasmids with ParR bound to the parC site (13). Growth of the stabilized filament forces the plasmids to the poles of the cell ensuring inheritance by both daughter cells.
The mechanism of action of Type I systems is less clear, although a mechanism similar to that observed with Type II systems has been suggested and is supported by some evidence. For example, several ParAs have been shown to undergo ATP-dependent polymerization in vitro (14–17). This raises the possibility that ParA mediates segregation in a manner similar to the actin-like ParM in that plasmids are either pushed or pulled apart by the dynamic assembly of the ATPase.
GFP fusions to at least four type I ParA ATPases (Soj, the chromosomal ParA homologue of B. subtilis, ParA of the E. coli plasmid pB171, SopA from F plasmid and ParAI from chromosome I of V. cholerae) have been shown to undergo movement within the cell (16, 18–22). This movement is similar to that observed with MinD, which oscillates between the ends of the cell in association with the membrane (23). Soj, SopA, and ParA from pB171 colocalize with the nucleoid and oscillate from one end of the nucleoid to the other or between nucleoids. ParAI from chromosome I of V. cholerae migrates across the cell from one end to the other (18, 19, 21, 22). Where examined, the ParB homologue is required for movement, whereas both nucleoid localization and oscillation require a functional ATPase (18, 19, 21). Because oscillation of ParA homologues and plasmid partitioning require the same components, it is possible that the oscillation of ParA homologues is important for plasmid partitioning and chromosome segregation processes.
Most plasmids and all known chromosomal loci encode Type I partitioning proteins. Replacing a plasmid par locus with one from the chromosome has been shown to stabilize plasmids indicating that the chromosomal par systems can function to mediate plasmid maintenance (5, 24–26). For example, the par locus from B. subtilis containing soj (parA), spo0J (parB), and parS can be used to stabilize a miniF deleted for its par locus (sopABC) (25).
Recent analysis of the chromosomal ParA homologue Soj from Thermus thermophilus revealed that it dimerizes and binds DNA in the presence of ATP (27). An ATPase-deficient mutant, Soj-D44A, crystallized as a dimer with ATP, similar to the NifH dimer and to that proposed for MinD (28, 29). The DNA binding observed in these studies is ATP-dependent, suggesting that Soj dimerized and then bound to DNA. Electron microscopy revealed that Soj bound to DNA formed a nucleoprotein filament, raising the possibility that a bound dimer recruited additional dimers to allow the spread of Soj on the DNA. This result indicated that Soj may be similar to MinD, which is thought to bind to the membrane as a dimer and further associate to undergo surface-dependent polymerization (30). The cooperative membrane binding along with MinE stimulation of the MinD ATPase underlie the mechanism that allows MinD to oscillate in the cell (31–33). Soj and Spo0J could be interacting in a similar manner. To explore the mechanism of Soj DNA binding and its role in Soj function, we sought to identify residues in Soj involved in DNA binding to determine whether this binding is required for Soj promoted plasmid segregation.
Results
GFP-Soj Disrupts Nucleoid Morphology in E. coli.
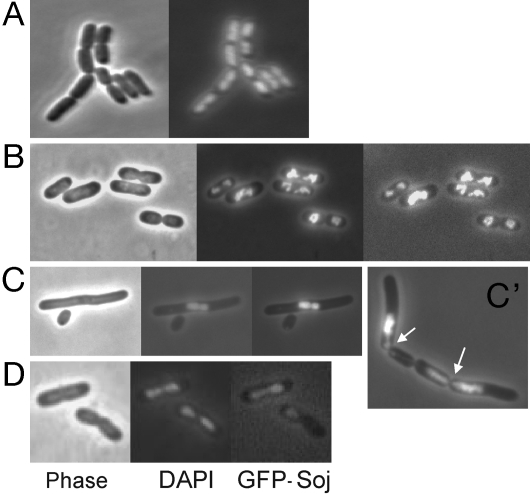
Expression of GFP-Soj in E. coli without Spo0J results in the accumulation of GFP-Soj on the nucleoid(s). Fig. 1B shows that GFP-Soj colocalizes with and distorts the nucleoid. Increased expression of GFP-Soj leads to further alterations in nucleoid morphology. After several hours of growth of W3110/pSEB200 (Plac::gfp-soj) in the presence of IPTG, the nucleoids were noticeably condensed, and the cells were elongated with extended nucleoid-free regions (Fig. 1C). For comparison, the nucleoid distribution in W3110 carrying pSEB181 (parent vector for pSEB200, encoding only GFP) is shown in Fig. 1A. In addition to the disruption of nucleoid morphology and segregation caused by overexpression of GFP-Soj, some anucleate cells were formed. Also, guillotining of the nucleoid occurred in a number of cells, indicating that the nucleoid occlusion system is no longer functioning properly (Fig. 1C′, arrows). The GFP-Soj always colocalized with the DNA and was not readily detected in the cytoplasm of cells containing DNA or in anucleate cells. The same effects on nucleoid morphology were also observed with His-tagged or untagged Soj indicating that they were not due to the GFP tag (data not shown).
Fig. 1.
GFP-Soj colocalizes with and distorts the nucleoid in E. coli in the absence of Spo0J. (A) W3110 cells stained with DAPI. (B) GFP-Soj expressed in W3110 from pSEB200. Cells shown are in early exponential phase, ≈1 h after induction with 50 μM IPTG. (C) GFP-Soj as in B except cells are in early stationary phase several hours after induction of GFP-Soj expression. (C′) Additional example showing that GFP-Soj caused nucleoid distortion, guillotining of nucleoids (arrows), filamentation and formation of anucleate cells. (D) Coexpression of Spo0J along with GFP-Soj (from pCMN003) protects cells from detrimental effects of GFP-Soj overexpression. Also, the GFP-Soj is asymmetrically localized in cells coexpressing Spo0J and oscillates. C′ is GFP fluorescence, but DAPI staining was identical.
When GFP-Soj was coexpressed with Spo0J in the context of the operon, GFP-Soj was frequently asymmetrically localized on the DNA (Fig. 1D). The GFP-Soj oscillated on the nucleoid in some of the cells on a time scale of minutes as observed in B. subtilis (data not shown). The presence of Spo0J and the parS site within spo0J was required for this movement (data not shown). The disruptive effects of Soj on nucleoid and cell morphologies were not observed if GFP-Soj was coexpressed with Spo0J (with or without parS), even though GFP-Soj colocalized with the nucleoids.
Identification of Conserved, Surface-Exposed Arginine Residues.
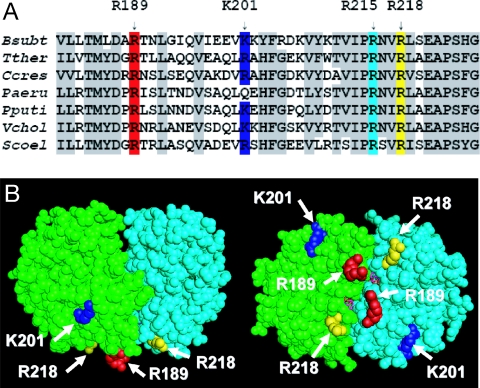
The solution of the T. thermophilus Soj crystal structure did not reveal any known structural DNA-binding motifs, although Leonard et al. (27) showed that the protein bound nonspecifically to DNA in an ATP-dependent manner. Because proteins that bind DNA nonspecifically often do so through positively charged residues that interact with the negatively charged phosphate backbone, we aligned Soj/ParA sequences from several bacterial species. We found two essentially invariant arginine residues that mapped to the surface of the T. thermophilus Soj structure, R189 and R218 (B. subtilis numbering) (Fig. 2A). These arginines are not conserved in MinD or plasmid ParAs. A third basic residue in the same region of the primary sequence, K201 (R194 in T. thermophilus), was also surface-exposed although it was not as conserved as R189 and R218. Another highly conserved arginine residue in this region, R215, is not completely exposed and is involved in binding ATP (27).
Fig. 2.
Identification of highly conserved, surface exposed basic residues. (A) A portion of an alignment of chromosomal ParA homologues is shown with conserved basic residues highlighted in color. Other conserved residues are shaded gray. (B) Conserved residues are highlighted on the dimer structure of Soj from T. thermophilus. The numbering is according to Soj from B. subtilis. R189 and R218 are near the dimer interface, whereas K201 (R194 of T. thermophilus Soj) sits back from the interface on the same face of the dimer. Bsubt, B. subtilis; Tther, T. thermophilus; Ccres, C. crescentus; Paeru, P. aeruginosa; Pputi, P. putida; Vchol, V. cholera; Scoel, S. coelicolor.
Fig. 2B shows the location of these residues on the Soj dimer structure. Both R189 and R218 sit on the same face of the dimer near the dimer interface. Although K201 is nearby, it is farther from the dimer interface.
Mutation of R189 and R218 Disrupts DNA Binding in Vivo.
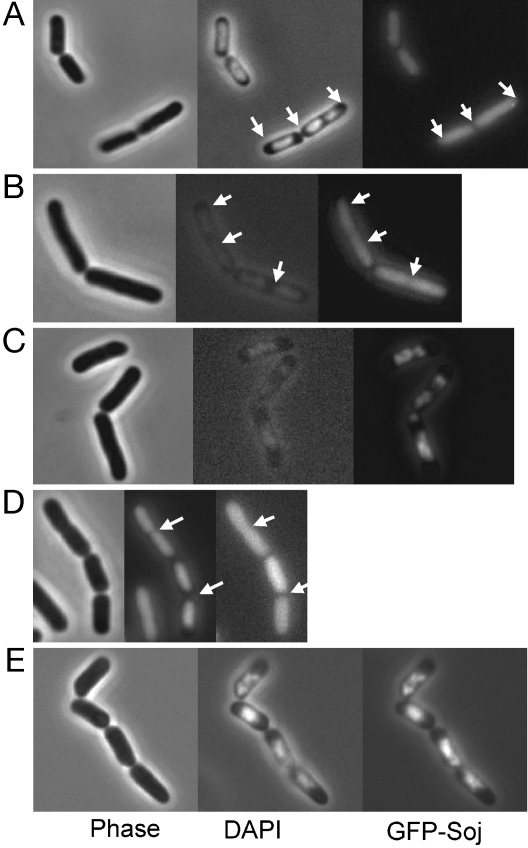
To determine whether the basic residues identified above were involved in the association of Soj with DNA, we exploited the phenotype induced by GFP-Soj expression in E. coli. Mutations were introduced into gfp-soj, and the effects of expression on cell morphology were analyzed. GFP-Soj-R189E and GFP-Soj-R218E did not affect nucleoid or cell morphology or colocalize with the DAPI stained nucleoid (Fig. 3 A and D). In contrast, GFP-Soj-K201E remained associated with the nucleoid and caused the morphological phenotypes observed with WT Soj (Fig. 3C).
Fig. 3.
Effect of mutation of the basic residues on GFP-Soj localization to the nucleoid. W3110 containing pSEB200 derivatives carrying various mutations were analyzed 2 hours after induction with IPTG. (A) R189E. (B) R189A. (C) K201E. (D) R218E. (E) R218A. Arrows indicate DNA-free regions occupied by the GFP-fusions.
The importance of these basic residues for DNA binding was further assessed by mutation to alanine. GFP-Soj-R189A did not colocalize with the nucleoid, and both the nucleoid and cells were normal in length and overall morphology (Fig. 3B). On the other hand, GFP-Soj-R218A associated with the nucleoid and induced morphological defects similar to WT Soj (Fig. 3E). In the presence of Spo0J, GFP-Soj-R218A was asymmetrically localized and displayed movement, similar to the WT. GFP-Soj-K201A also behaved like WT (data not shown). All mutant proteins were stable and expressed to approximately the same level as WT Soj [supporting information (SI) Fig. 8]. Together, these results indicate that residues R189 and R218 are involved in mediating the interaction of Soj with the nucleoid, whereas K201 is not involved.
Mutations Do Not Affect ATP-Dependent Dimerization.
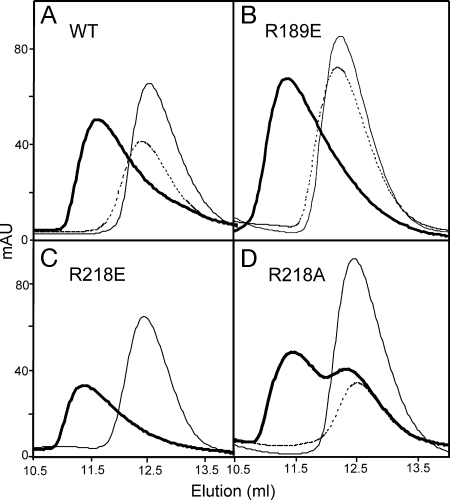
To verify that mutation of the arginine residues had no effect on ATP-dependent dimerization, we purified the WT and mutant proteins fused to an N-terminal histidine tag. Analysis by size-exclusion chromatography revealed they all migrated as monomers when run with ADP or no nucleotide (Fig. 4A–D; R218E without nucleotide was not run). When run with ATP, the proteins eluted as dimers, indicating that R189 and R218 could be substituted with alanine or glutamate without affecting dimerization. We did note that, although R218A dimerizes in the presence of ATP, a fraction of this protein eluted as a monomer (Fig. 4D).
Fig. 4.
Analysis of mutating basic residues on the dimerization of Soj. Size-exclusion chromatography of purified WT Soj and Soj mutant proteins (0.2 mM). Dashed lines, no nucleotide; thick lines, ATP (0.2 mM); thin lines, ADP (0.2 mM). Elution of size standards: cytochrome C (12.4K), 14.3 ml; carbonic anhydrase (29K), 12.4 ml; and BSA (66K), 10.3 ml. Absorbance was monitored at 280 nm.
Mutations Disrupt DNA Binding in Vitro.
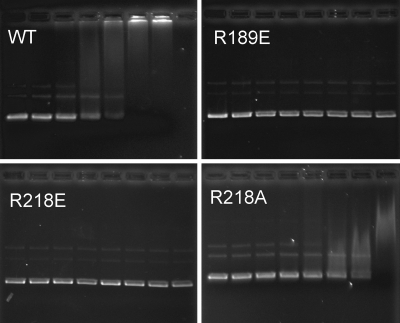
To test for effects on DNA binding, the His-tagged Soj mutant proteins were used in an electrophoretic mobility shift assay with pUC18 (2.7 kb). No shift in the migration of the DNA was detected with WT Soj or the mutants in the presence of ADP or no nucleotide (data not shown). In the presence of ATP WT Soj prevented DNA from entering the gel at concentrations >7.5 μM (Fig. 5). The abrupt transition suggests cooperativity in the binding of Soj to the DNA as also observed by Leonard et al. (27). In contrast, the two mutant proteins that do not localize to the nucleoid in vivo, R189E and R218E, had no effect on the migration of the DNA, even at 25 μM (Fig. 5, rightmost lane). Consistent with its in vivo localization, R218A still bound DNA, but a higher concentration of protein was required to produce a shift. At 10–12.5 μM R218A, the DNA starts to shift, but a complete shift of the DNA is observed only at 25 μM (Fig. 5). This result indicates that R218A binds to DNA but has a reduced affinity compared with WT Soj. This reduced affinity could be the result of the less efficient dimerization observed during the size-exclusion chromatography (Fig. 4D). However, it is likely due to loss of the positive charge on the dimer surface, because a negative charge at this position (R218E) did not affect dimerization even though DNA binding was absent.
Fig. 5.
Electrophoretic mobility shift assay of DNA binding by WT Soj and Soj mutant proteins. Increasing amounts of WT or mutant Soj proteins were incubated with pUC18 DNA (12.6 nM) in the presence of either ADP or ATP and run on an agarose gel. Protein concentrations for each set of reactions were as follows: Lane 1, 0 μM Soj; lane 2, 1.25 μM; lane 3, 2.5 μM; lane 4, 5 μM; lane 5, 7.5 μM; lane 6, 10 μM; lane 7, 12.5 μM; and lane 8, 25 μM (not done for WT). Only results with ATP are shown. No shift was observed with ADP.
Soj DNA Binding Is Required for Stabilization of miniF by soj spo0J parS.
Yamaichi and Niki (25) demonstrated that B. subtilis Soj and Spo0J along with at least one copy of parS could promote the stability of an unstable miniF plasmid. To determine whether or not the nonspecific DNA binding activity of Soj is important for its role in plasmid partitioning, we replaced the WT soj allele in the test plasmid pXX765 with R189E and R218A. This plasmid is a miniF plasmid containing the B. subtilis soj locus as the only functional partitioning locus and was used by Yamaichi and Niki (25).
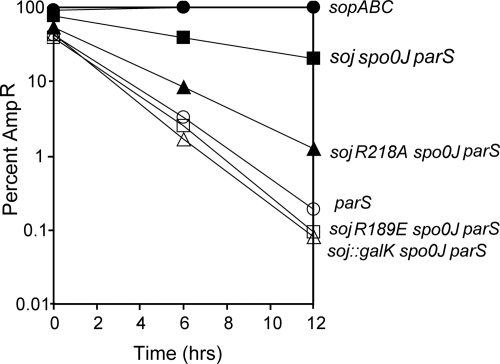
To test for stability, JS238 carrying the plasmids was grown with selection, diluted into media without antibiotic, and maintained in exponential growth for 12 h, or ≈24 generations (Fig. 6). After 12 h, pXX765 (WT soj, spo0J, and parS) was present in ≈20% of the population, consistent with the previously reported stabilization of this plasmid in E. coli (25). In contrast, pCMN033 carrying Soj-R189E substituted for the WT soj allele was essentially lost from the population, as were control plasmids with only parS (pXX764) or in which soj was disrupted by insertion of galK (pCMN032). pCMN033 (R218A) displayed an intermediate stability and was retained by ≈1% of the population after 12 h of unselected growth.
Fig. 6.
Effect of soj mutations on plasmid stability. The stability of miniF plasmids bearing WT or mutant soj alleles was determined. The percentage of plasmid-bearing cells was determined for each strain after dilution into nonselective medium and growth for 0, 6 and 12 h. The average values from three experiments were plotted for pXX765, pCMN033 (R189E), pCMN033 (R218A), and pXX764, whereas the average values from two experiments were plotted for pXX704 and pCMN032.
The plasmid carrying WT soj (pXX765) was lost at a rate of 5% per generation, whereas pCMN033 (R218A) was lost at a rate of 14% per generation. pCMN033 (R189E) was lost at a rate of 20% per generation, as were the control plasmids pXX764 (parS only; 22% per generation) and pCMN032 (soj::galK spo0J parS; 23% per generation). It has been shown that the soj and spo0J genes do not affect copy number or multimer resolution of the miniF plasmid (25). The results obtained with the above mutants indicate that DNA binding by Soj is essential to promote plasmid stability.
Discussion
ParA proteins play a critical role in plasmid and chromosome segregation, although the mechanism of Type I ParAs is not clear. In this study, we examined a ParA homologue from B. subtilis, which, along with Spo0J and parS, can stabilize a plasmid in E. coli. We found that DNA binding by Soj is essential for plasmid segregation.
DNA binding by Soj has been described; however, the region(s) of the protein responsible had not been identified. Because nonspecific DNA binding usually involves basic residues, we looked for conserved arginine residues and assessed their role by examining the effects of altering these residues on the ability of Soj to condense the nucleoid in vivo and bind DNA in vitro. Our analysis points to the importance of arginine residues that lie on one face of the Soj dimer. One of these arginines, R189, appears to be essential for binding, because changing it to alanine or glutamic acid eliminated DNA binding. Arginine at position 218 also plays a role because changing it to glutamate eliminated binding and changing it to alanine reduced but did not eliminate the ability of Soj to bind DNA.
Although it was observed that Soj colocalizes with the nucleoid, a role for DNA binding in the function of this protein was unclear. We have shown here that mutations that prevent Soj from localizing to the nucleoid also prevent it from binding DNA and supporting plasmid segregation. Notably, the degree to which the DNA binding is affected correlates with the severity of the effect on plasmid stabilization. The R189E mutation eliminated DNA binding and plasmid stabilization (≈100-fold reduction), whereas the R218A mutation reduced the affinity for DNA and reduced plasmid stability (≈10-fold reduction).
Condensation of the nucleoid by Soj is due to its ability to bind to DNA and is a convenient in vivo assay for DNA binding. Nucleoid condensation is also observed upon overexpression of other nonspecific DNA-binding proteins (34). Interestingly, the condensation of the DNA by Soj resulted in guillotining of the nucleoid, indicating that nucleoid occlusion (NOC), which prevents septation from occurring over nucleoids (35, 36), was suppressed. This could be the result of Soj competing with effectors of NOC [SlmA in E. coli (35)] for binding to the nucleoid or to the altered nucleoid structure interfering with NOC function. This effect of Soj was ameliorated by the presence of Spo0J, suggesting that Spo0J antagonizes this effect of Soj.
Model for Soj Binding to DNA.
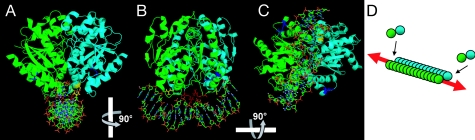
In this and previous work (27), it was shown that ATP is required for dimerization of Soj and for it to bind double-stranded DNA. Our findings indicate that the DNA-binding site on the Soj surface includes conserved arginines. Based on these findings, it was possible to envision the Soj dimer binding to DNA. The approximate distances between R189 and R218 are similar to the distances between the phosphate backbone from the inside edge of the major groove to the nearest edge of the minor groove. By using this information and the DNA structure from the Protein Data Bank file 1SRS, a manually docked model of Soj bound to DNA was created in DeepView/Swiss-PdbViewer. This manually docked model was subjected to energy minimization to account for steric hindrances, and the model depicted in Fig. 7A–C was generated (details in SI Text).
Fig. 7.
Model for binding of Soj to DNA. (A) The docked model was created as described in SI Text by using chains A and B from the Soj dimer structure (2BEK) and the DNA from the SRF core complex (1SRS). The model in A was rotated to yield the views in B and C. (D) Model for polymerization of Soj dimers on DNA.
In this model, the key role of R189 is clear. Upon dimerization, the two R189 residues are appropriately positioned to make contact with the phosphate backbones of the minor groove, potentially stabilizing the interaction of Soj with the DNA. In this model, R218 interacts with the phosphate backbone of the major groove on either side of the dimer, thus illustrating how this residue could also play a role in binding to the DNA. The interaction of Soj with the DNA is likely stabilized by other undetermined residues as well. Note that K201, which is not required for DNA binding, does not come into contact with the DNA in this model. A bend in the DNA was required for R189 and R218 to be in close proximity to the phosphate backbone. The condensation of the DNA observed within E. coli cells overexpressing Soj could be the result of the cumulative effects of Soj-induced bending.
As shown in Fig. 7 A–C, the dimer interface is parallel to the DNA in our model. Our docked model also illustrates how Soj DNA binding could be cooperative. Binding by one Soj dimer provides the next incoming dimer with two sites for interaction: with the first Soj dimer and with the DNA. Also, this orientation of the Soj dimer on the DNA predicts that the dimer interface is parallel to the axis of polymerization (Fig. 7D). Importantly, polymerization of Soj would lack polarity, because each side of the dimer along the DNA axis presents an equivalent polymerization interface. It will be interesting to determine whether MinD also assembles in a similar manner.
DNA Binding by Soj and Other ParAs.
Although we studied Soj and the role of DNA binding in plasmid segregation, several observations suggest that nonspecific DNA binding is an important property of all chromosomally encoded ParA proteins that also extends to plasmid ParAs. First of all, the two arginine residues we identified here are conserved in chromosomally encoded ParAs. Such sequence conservation implies that their function is also conserved. The only other chromosomal ParA to be examined for DNA binding is ParA of C. crescentus (37). Although it was observed to bind single-stranded DNA, binding to double-stranded DNA was not detected. However, the highest concentration of ParA tested was <1 μM, too low for binding to double-stranded DNA to have been observed.
The localization of several plasmid ParA proteins has been examined after fusion to GFP. The most extensively studied is ParA of pB171, which, like Soj, oscillates on the nucleoid. Deconvolution of images of the oscillation suggests that the ParA is in spiral structures (18). Mutations in the Walker A motif prevented spiral formation, suggesting that ATP is required for their formation. We have purified this ParA and found that it also binds nonspecifically to DNA in vitro in an ATP-dependent manner similar to Soj (data not shown). This ParA is a type Ib that lacks the N-terminal extension involved in specific DNA binding observed with type Ia ParAs. A type Ia ParA, SopA of the F plasmid, is nucleoid associated in the absence of SopB and oscillates on nucleoids when SopB and sopC are present (16). It also binds DNA nonspecifically in vitro although that aspect of its behavior has not been explored thoroughly (17). We would suggest that this nonspecific DNA binding does not require the N-terminal extension that is likely involved in the specific binding required for regulation of the sop operon.
There are substantial differences in the primary amino acid sequences within the ParA superfamily. There is 21% amino acid identity between B. subtilis Soj and ParA from pB171, and there is 21% amino acid identity between Soj and SopA. Based on extensive alignments, it has not been possible to identify conserved arginine residues that may be important for nonspecific DNA binding by SopA or ParA. In the absence of structural data, empirical evidence will be necessary to determine which residues are involved in nonspecific DNA binding, however, it is reasonable to suggest that the binding surface will be generated upon dimerization, as it is for Soj.
Assembly of Proteins of the ParA Family.
Several studies have demonstrated that ParA proteins can polymerize in vitro independently of DNA (14–17). ParF from plasmid TP228, ParA from pB171, and SopA from F have been shown to polymerize by light scattering, sedimentation, and electron microscopy (14–17). The electron microscopy reveals small bundles of polymers similar to those reported in one study of MinD (38). However, several results suggest some caution in interpreting the physiological relevance of these bundles. For instance, in another study of MinD, polymers were observed only on the surface of phospholipid vesicles (30). In the absence of vesicles, MinD dimerized, but higher-order complexes were not observed (39). Dimerization, but not further assembly, was also observed with Soj from T. thermophilus in the absence of DNA (27). When DNA was present, Soj bound cooperatively to generate nucleoprotein filaments.
Bouet et al. (17) reported that SopA polymerized in an ATP-dependent fashion. The addition of double-stranded DNA, but not single-stranded DNA, prevented the formation of polymers. Sequestering the DNA with another DNA-binding protein, such as SopB, which also binds DNA nonspecifically, restored SopA polymerization. The authors suggested that SopA is stored on DNA to prevent inappropriate polymerization and that, in an area of the cell where the DNA is masked, perhaps by SopB, the displaced SopA is free to form filaments for the partitioning reaction. Our results suggest, at least for Soj, that the assembly on the DNA is the essential function in partitioning.
The polymerization and bundling of ParA homologues that has been observed in the absence of DNA may simply reflect their tendency to self-associate to form dimers and higher-order oligomers in the presence of ATP. Perhaps the filament bundling observed in the absence of DNA, and presumably responsible for the light scattering and sedimentation, does not occur if DNA is present, because binding to DNA may mask the bundling surface. In the cell, chromosomal and plasmid DNA are available to serve as substrates for assembly of these proteins, questioning whether polymers would form independently of DNA in vivo.
Plasmid Stabilization.
In models of plasmid segregation it has been suggested that ParA is actively involved in plasmid segregation and that polymerization of ParA mediates plasmid movement. Fogel and Waldor (22) observed that after duplication of the origin and the nearby ParB-parS focus, one ParB-parS focus and associated origin remained near the pole, whereas the other followed a cloud of ParAI as it moved to the other pole, resulting in segregation of the associated origin. They proposed that the cloud represented ParAI filaments and that the ParB-parS complex induces the depolymerization of these filaments, resulting in the movement of the ParB complex by a “Brownian ratchet” mechanism similar to the MinE ring chasing MinD off the membrane. The only change we would suggest is that this cloud of ParAI is ParAI polymerized on the DNA rather than freely polymerized ParAI. Leonard et al. (40) have proposed a similar model. In summary, we have identified the region of Soj involved in DNA binding, provided a model for this binding, and provided evidence that this property of a ParA homologue is critical for plasmid stabilization.
Experimental Procedures
Bacterial Strains and Plasmids.
Strains used in this work were E. coli K12 strains W3110 and JS238 (41). Plasmids used for testing segregation were derivatives of pXX765, which is a miniF plasmid containing B. subtilis soj spo0J and two parS sites (25). Derivatives were made carrying R189E or R218A as described in detail in the SI Text. pSEB200 (Plac::gfp-soj) and derivatives containing soj mutations were used to examine GFP-Soj localization. A complete listing is in SI Table 1.
Analysis of GFP-Soj Localization.
For analysis of the localization of GFP-Soj, overnight cultures of W3110 bearing either pSEB200 (Plac::gfp-soj) or pCMN003 (Plac::gfp-soj spo0J) were diluted in LB for 2 h, induced with IPTG (50–100 μM), and maintained in exponential phase by dilution or allowed to enter stationary phase. For the mutant GFP-Soj fusions, W3110 bearing the appropriate plasmids was streaked directly onto an LB plate supplemented with spectinomycin and 500 μM IPTG and incubated at 37°C for 2–6 h. Cells were stained with DAPI and mounted on glass slides. Microscopy was performed as described (42).
Determining Plasmid Stability.
Plasmid-stability assays were performed as described in Yamaichi and Niki (25) with minor modifications. Strains were grown to exponential phase under selection, transferred to media without selection, and maintained in exponential growth by dilution. Cultures were sampled at the times indicated to determine the viable count and number of cells containing the plasmid. The percentage of plasmids lost per generation (L) was determined by using the equation L = [1-(Ff/Fi)1/n] × 100, where Fi is the fraction of cells initially carrying the plasmid, and Ff is the fraction of plasmid bearing cells after n generations of nonselective growth (43). Values were determined by using the values plotted in Fig. 6.
DNA Binding Assay.
Electrophoretic mobility shift assays were performed as described (27). We used pUC18 plasmid (2.7 kb) at a final concentration of 12.6 nM. Reaction components (except Soj) were dispensed into individual reaction tubes from a concentrated master mix. Soj Dilution Buffer (50 mM Tris, pH 7.5, 100 mM NaCl, 1 mM DTT, 10% glycerol) was added as needed to give a final reaction volume of 10 μl, and then Soj was added to yield the final concentrations indicated. Reactions were incubated for 10 min at room temperature before being run on a 1% agarose gel as described by Leonard et al. (27).
Supplementary Material
ACKNOWLEDGMENTS.
We thank Todd Holyoak for assistance with modeling of the Soj–DNA complex and Nicholas Bartelli for assistance in repeating some experiments. We also thank H. Niki of the National Institute of Genetics (Japan) for providing the miniF constructs. This work was supported by National Institutes of Health Grant GM27964.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705196105/DC1.
References
- 1.Ebersbach G, Gerdes K. Annu Rev Genet. 2005;39:453–479. doi: 10.1146/annurev.genet.38.072902.091252. [DOI] [PubMed] [Google Scholar]
- 2.Ghosh SK, Hajra S, Paek A, Jayaram M. Annu Rev Biochem. 2006;75:211–241. doi: 10.1146/annurev.biochem.75.101304.124037. [DOI] [PubMed] [Google Scholar]
- 3.Mohl DA, Gober JW. Cell. 1997;88:675–684. doi: 10.1016/s0092-8674(00)81910-8. [DOI] [PubMed] [Google Scholar]
- 4.Lewis RA, Bignell CR, Zeng W, Jones AC, Thomas CM. Microbiology. 2002;148:537–548. doi: 10.1099/00221287-148-2-537. [DOI] [PubMed] [Google Scholar]
- 5.Bartosik AA, Lasocki K, Mierzejewska J, Thomas CM, Jagura-Burdzy G. J Bacteriol. 2004;186:6983–6998. doi: 10.1128/JB.186.20.6983-6998.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim HJ, Calcutt MJ, Schmidt FJ, Chater KF. J Bacteriol. 2000;182:1313–1320. doi: 10.1128/jb.182.5.1313-1320.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saint-Dic D, Frushour BP, Kehrl JH, Kahng LS. J Bacteriol. 2006;188:5626–5631. doi: 10.1128/JB.00250-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ireton K, Gunther NW, Grossman AD. J Bacteriol. 1994;176:5320–5329. doi: 10.1128/jb.176.17.5320-5329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerdes K, Moller-Jensen J, Bugge Jensen R. Mol Microbiol. 2000;37:455–466. doi: 10.1046/j.1365-2958.2000.01975.x. [DOI] [PubMed] [Google Scholar]
- 10.Gerdes K, Moller-Jensen J, Ebersbach G, Kruse T, Nordstrom K. Cell. 2004;116:359–366. doi: 10.1016/s0092-8674(04)00116-3. [DOI] [PubMed] [Google Scholar]
- 11.Moller-Jensen J, Jensen RB, Lowe J, Gerdes K. EMBO J. 2002;21:3119–3127. doi: 10.1093/emboj/cdf320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moller-Jensen J, Borch J, Dam M, Jensen RB, Roepstorff P, Gerdes K. Mol Cell. 2003;12:1477–1487. doi: 10.1016/s1097-2765(03)00451-9. [DOI] [PubMed] [Google Scholar]
- 13.Garner EC, Campbell CS, Mullins RD. Science. 2004;306:1021–1025. doi: 10.1126/science.1101313. [DOI] [PubMed] [Google Scholar]
- 14.Ebersbach G, Ringgaard S, Moller-Jensen J, Wang Q, Sherratt DJ, Gerdes K. Mol Microbiol. 2006;61:1428–1442. doi: 10.1111/j.1365-2958.2006.05322.x. [DOI] [PubMed] [Google Scholar]
- 15.Barilla D, Rosenberg MF, Nobbmann U, Hayes F. EMBO J. 2005;24:1453–1464. doi: 10.1038/sj.emboj.7600619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim GE, Derman AI, Pogliano J. Proc Natl Acad Sci USA. 2005;102:17658–17663. doi: 10.1073/pnas.0507222102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bouet JY, Ah-Seng Y, Benmeradi N, Lane D. Mol Microbiol. 2007;63:468–481. doi: 10.1111/j.1365-2958.2006.05537.x. [DOI] [PubMed] [Google Scholar]
- 18.Quisel JD, Lin DC, Grossman AD. Mol Cell. 1999;4:665–672. doi: 10.1016/s1097-2765(00)80377-9. [DOI] [PubMed] [Google Scholar]
- 19.Marston AL, Errington J. Mol Cell. 1999;4:673–682. doi: 10.1016/s1097-2765(00)80378-0. [DOI] [PubMed] [Google Scholar]
- 20.Ebersbach G, Gerdes K. Mol Microbiol. 2004;52:385–398. doi: 10.1111/j.1365-2958.2004.04002.x. [DOI] [PubMed] [Google Scholar]
- 21.Ebersbach G, Gerdes K. Proc Natl Acad Sci USA. 2001;98:15078–15083. doi: 10.1073/pnas.261569598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fogel MA, Waldor MK. Genes Dev. 2006;20:3269–3282. doi: 10.1101/gad.1496506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raskin DM, de Boer PA. Proc Natl Acad Sci USA. 1999;96:4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Godfrin-Estevenon AM, Pasta F, Lane D. Mol Microbiol. 2002;43:39–49. doi: 10.1046/j.1365-2958.2002.02735.x. [DOI] [PubMed] [Google Scholar]
- 25.Yamaichi Y, Niki H. Proc Natl Acad Sci USA. 2000;97:14656–14661. doi: 10.1073/pnas.97.26.14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dubarry N, Pasta F, Lane D. J Bacteriol. 2006;188:1489–1496. doi: 10.1128/JB.188.4.1489-1496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leonard TA, Butler PJ, Lowe J. EMBO J. 2005;24:270–282. doi: 10.1038/sj.emboj.7600530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Georgiadis MM, Komiya H, Chakrabarti P, Woo D, Kornuc JJ, Rees DC. Science. 1992;257:1653–1659. doi: 10.1126/science.1529353. [DOI] [PubMed] [Google Scholar]
- 29.Lutkenhaus J, Sundaramoorthy M. Mol Microbiol. 2003;48:295–303. doi: 10.1046/j.1365-2958.2003.03427.x. [DOI] [PubMed] [Google Scholar]
- 30.Hu Z, Gogol EP, Lutkenhaus J. Proc Natl Acad Sci USA. 2002;99:6761–6766. doi: 10.1073/pnas.102059099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu Z, Lutkenhaus J. Mol Cell. 2001;7:1337–1343. doi: 10.1016/s1097-2765(01)00273-8. [DOI] [PubMed] [Google Scholar]
- 32.Lackner LL, Raskin DM, de Boer PA. J Bacteriol. 2003;185:735–749. doi: 10.1128/JB.185.3.735-749.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mileykovskaya E, Fishov I, Fu X, Corbin BD, Margolin W, Dowhan W. J Biol Chem. 2003;278:22193–22198. doi: 10.1074/jbc.M302603200. [DOI] [PubMed] [Google Scholar]
- 34.Setlow B, Hand AR, Setlow P. J Bacteriol. 1991;173:1642–1653. doi: 10.1128/jb.173.5.1642-1653.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernhardt TG, de Boer PAJ. Mol Cell. 2005;18:555–564. doi: 10.1016/j.molcel.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu LJ, Errington J. Cell. 2004;117:915–925. doi: 10.1016/j.cell.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 37.Easter J, Jr, Gober JW. Mol Cell. 2002;10:427–434. doi: 10.1016/s1097-2765(02)00594-4. [DOI] [PubMed] [Google Scholar]
- 38.Suefuji K, Valluzzi R, RayChaudhuri D. Proc Natl Acad Sci USA. 2002;99:16776–16781. doi: 10.1073/pnas.262671699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu Z, Saez C, Lutkenhaus J. J Bacteriol. 2003;185:196–203. doi: 10.1128/JB.185.1.196-203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leonard TA, Moller-Jensen J, Lowe J. Philos Trans R Soc London Ser B. 2005;360:523–535. doi: 10.1098/rstb.2004.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pichoff S, Vollrath B, Touriol C, Bouche JP. Mol Microbiol. 1995;18:321–329. doi: 10.1111/j.1365-2958.1995.mmi_18020321.x. [DOI] [PubMed] [Google Scholar]
- 42.Pichoff S, Lutkenhaus J. Mol Microbiol. 2005;55:1722–1734. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- 43.Ravin N, Lane D. J Bacteriol. 1999;181:6898–6906. doi: 10.1128/jb.181.22.6898-6906.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.