Abstract
Circadian rhythms help organisms adapt to predictable daily changes in their environment. Light resets the phase of the underlying oscillator to maintain the organism in sync with its surroundings. Light also affects the amplitude of overt rhythms. At a critical phase during the night, when phase shifts are maximal, light can reduce rhythm amplitude to nearly zero, whereas in the subjective day, when phase shifts are minimal, it can boost amplitude substantially. To explore the cellular basis for this reciprocal relationship between phase shift and amplitude change, we generated a photoentrainable, cell-based system in mammalian fibroblasts that shares several key features of suprachiasmatic nucleus light entrainment. Upon light stimulation, these cells exhibit calcium/cyclic AMP responsive element-binding (CREB) protein phosphorylation, leading to temporally gated acute induction of the Per2 gene, followed by phase-dependent changes in phase and/or amplitude of the PER2 circadian rhythm. At phases near the PER2 peak, photic stimulation causes little phase shift but enhanced rhythm amplitude. At phases near the PER2 nadir, on the other hand, the same stimuli cause large phase shifts but dampen rhythm amplitude. Real-time monitoring of PER2 oscillations in single cells reveals that changes in both synchrony and amplitude of individual oscillators underlie these phenomena.
Keywords: circadian rhythm, melanopsin, singularity
Circadian oscillators enable organisms to anticipate and synchronize their behavior and physiology to periodic changes in the environment. In mammals, the hypothalamic suprachiasmatic nucleus (SCN) functions as a master circadian pacemaker, coordinating tissue-autonomous oscillators to generate overt rhythms (1, 2). At the molecular level, circadian rhythms are generated by a transcription–translation feedback loop. In this circuit, the transcriptional activators CLOCK and BMAL1 drive expression of the Cryptochrome 1 (Cry1), Cry2, Period 1 (Per1) and Per2 genes, whose protein products, in turn, repress CLOCK/BMAL1 transcriptional activity (reviewed in ref. 3). The phase of rhythms in mRNA and protein levels of these repressors, particularly Per2, reflects the phase of the oscillator (4).
Light entrains the SCN pacemaker, which relays phase information to peripheral oscillators via humoral and synaptic mechanisms. The retinorecipient cells of the SCN receive direct synaptic input from the intrinsically photosensitive retinal ganglion cells (ipRGCs) that express melanopsin. Upon photostimulation, the ipRGCs release neurotransmitters, which act via their cognate receptors to phosphorylate the calcium/cAMP-responsive element-binding protein (CREB). In turn, transcriptionally active phospho-CREB (pCREB) binds to Per1 and Per2 promoter CRE sites and activates transcription, subsequently resetting the phase of the molecular oscillator (reviewed in refs. 5 and 6). Mice bearing a targeted mutation in the CREB phosphorylation site (7) or perturbed Per function (8, 9) exhibit attenuated phase resetting.
The responses of the clock to entrainment cues are gated in a phase-specific manner. Light during the subjective day triggers little Per gene induction and minimal phase shifts, whereas an identical stimulus during the subjective night induces Per transcription and resets the oscillator (10, 11). Comparable gating is also observed in light-induced electrical responses of SCN neurons (12). Interestingly, in mammals as well as in other organisms, there is a “critical phase” in mid-subjective night when light reduces the amplitude of overt rhythms so severely that arrhythmicity or “singularity” results (13–17). Conversely, amplitude increase by light pulses at certain circadian phases is also well documented (18, 19). Under natural seasonal changes in day-lengths, these phenomena may encode photoperiod information in the amplitudes and phases of the SCN oscillators (20, 21). Existence of this singularity phenomenon in organisms from bacteria to mammals suggests that the mechanism is cell-autonomous. Two mechanisms have been suggested to explain overt amplitude reductions: (i) amplitude damping or (ii) desynchronization of individual oscillators (13, 22). These hypotheses have never been conclusively tested because of technical challenges and lack of an appropriate cellular system. To test these hypotheses, we generated a light-entrainable, cell-based system in mouse fibroblasts, where phase shifts and amplitude changes can be longitudinally monitored at both cell population and single-cell levels.
Mammalian fibroblasts possess cell-autonomous circadian oscillators (23) that share key properties with SCN neurons (2, 24). However, because medium change alone can reset the fibroblast clock (25, 26), it is not possible to use pharmacological agents to precisely manipulate oscillator resetting. Thus, to establish a suitable cellular system, we stably expressed melanopsin in mouse fibroblasts derived from the Per2Luciferase (Per2Luc) knockin mouse (2). In this system, light can be used as a phase-resetting agent and PER2::LUC bioluminescence levels as an oscillator phase reporter. Light pulses delivered at the peak level of the PER2::LUC bioluminescence rhythm (CT12) produced no phase shifts, but enhanced its amplitude in subsequent days. Similar light pulses at the opposite phase (CT0) produced larger phase shifts but severely damped amplitude. Single-cell bioluminescence measurements revealed that light at CT0 causes both phase desynchronization and amplitude reduction of individual cells, leading to singularity. However, similar light stimulation at CT12 synchronizes phases and thereby improves the aggregate amplitude. These results clearly show a reciprocal relationship between amplitude and phase-shift magnitude in clock resetting, which has practical implications for human circadian phase shifts.
Results
Light-Induced Phase Shifts in Melanopsin-Expressing Fibroblasts.
To establish a light-entrainable oscillator, we stably expressed mouse melanopsin (mOpn4) in immortalized fibroblasts derived from the Per2Luc knockin mouse (2). Clonal cell lines that stably express mouse melanopsin (Per2Luc;mOpn4) exhibited characteristic melanopsin photoresponses (5) [supporting information (SI) Fig. 6]. Both Per2Luc and Per2Luc;mOpn4 cell lines exhibited ≈24-h bioluminescence rhythms that persisted for at least 5 days (SI Fig. 6) with no significant difference in period length.
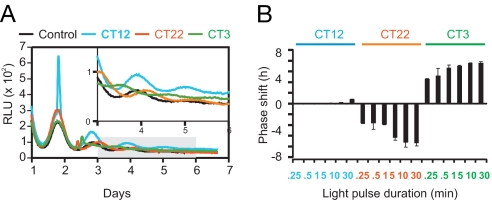
To determine whether light can phase-shift the Per2Luc;mOpn4 cells, culture dishes were either dark- or light-treated at three different phases: circadian times (CT) 12, 22, and 3, where CT12 corresponds to the peak phase of PER2::LUC rhythms. Light pulses at CT12 produced large transient increases in PER2::LUC levels but had little effect on the phase of the rhythm (Fig. 1A). In contrast, light pulses at CT22 and CT3 produced much smaller PER2::LUC inductions but substantial phase delays and advances, respectively. Per2Luc;mOpn4 cells are highly photosensitive, with as little as 15 sec of light at CT22 and CT3 producing phase shifts of ≈4 h. The phase-shift magnitude increased with light-stimulus duration, saturating at 10 min (Fig. 1B), which was the condition used for the rest of the experiments.
Fig. 1.
Light-induced phase shifts of PER2::LUC rhythms in Per2Luc;mOpn4 cells. (A) Per2Luc;mOpn4 cells exhibit rhythmic bioluminescence that can be reset by light. Representative examples of bioluminescence rhythms [relative light units/min (RLU)] and phase shifts in response to 10-min light pulses are shown. Shaded area is enlarged in the Inset. (B) Magnitude of phase shift [mean ± SD (n = 3)] increases with increasing duration of light pulse (3 × 1015 photons cm−2 s−1, 480 nm) at CT3 and CT22 but not at CT12 (CT12 = peak PER2::LUC level).
Photostimulation of Per2Luc;mOpn4 Cells Triggers pCREB-Mediated Transient Induction of Per2.
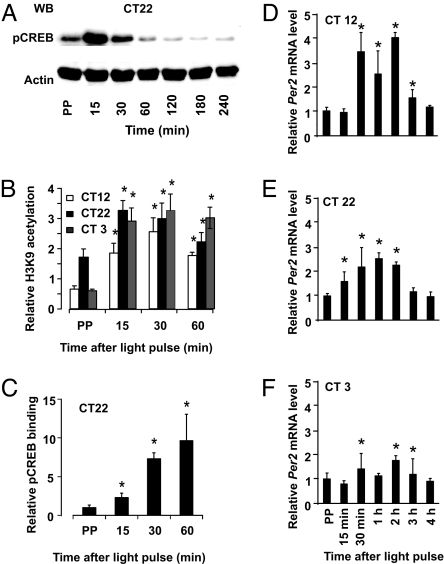
We tested whether light-activated Per gene induction in the Per2Luc;mOpn4 cells involves CREB phosphorylation. Immunoblot analysis of total-cell lysates revealed that pSer-133 CREB level increased within 15 min of light stimulation at CT22 (Fig. 2A), CT12, and CT3 (data not shown), which parallels a similar event in the SCN (27).
Fig. 2.
Light-induced CREB phosphorylation and Per gene transcription in Per2Luc;mOpn4 cells. (A) Immunoblot analysis shows light-induced rise in pCREB levels after light stimulation at CT22. (B and C) Chromatin immunoprecipitation with antiacetylated H3K9 (B) and anti-pSer-133 CREB antibody (C) indicate enrichment of immunoprecipitated Per2 promoter DNA sequences relative to the antibody control in response to light pulses at CT12, CT22, or CT3 for acetylated H3K9 and at CT22 for pCREB. (D–F) q-PCR analysis shows that light exposure at CT12 (D), CT22 (E), or CT3 (F) increased Per2 mRNA levels compared with dark-treated samples. Samples were collected before light pulse (prepulse, or PP) as well as at indicated times (x axis) after 10-min light stimulation. Values in B–F are mean ± SEM (B and C) or mean ± SD (D–F); n = 3; *, P < 0.01 compared with the prepulse; Student's t test.
Transcriptionally active pCREB binds to cis-acting CRE sites with favorable chromatin modifications. Because Histone H3 acetylation at Lys-9 (H3K9) correlates with transcriptional activation (28), we tested the extent of H3K9 acetylation and pCREB occupancy at the Per2 promoter CRE site by ChIP assays. The levels of H3K9 acetylated histones at the Per2 CRE site increased significantly within 15 min after photostimulation, closely followed by increased pCREB binding (Fig. 2 B and C). Subsequently, Per2 mRNA began to increase, peaking 30–60 min after a light pulse and declining afterward (Fig. 2 D–F). Similar results were obtained for Per1 transcription. As seen with PER2::LUC induction, the increase in Per2 mRNA relative to its prelight pulse level was remarkably higher at CT12 than at CT22 and CT3.
Circadian Modulation of Per Induction.
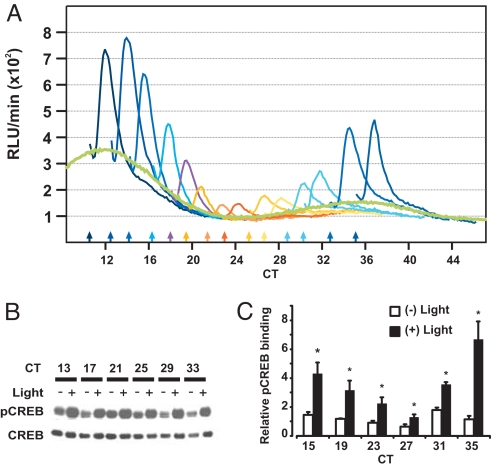
Next, we systematically presented 10-min light pulses to Per2Luc;mOpn4 cells at regular intervals over a 24-h period. As observed earlier (Fig. 1A), the magnitude of acute induction of PER2::LUC exhibited a clear circadian modulation, with the largest induction at approximately CT12 and minimal induction at approximately CT0 (Fig. 3A). A similar photic gating phenomenon in the SCN correlates with the gating of CREB phosphorylation (27).
Fig. 3.
Circadian modulation of PER2::LUC induction correlates with pCREB binding to the Per2 promoter. (A) Median normalized bioluminescence counts (thin colored lines) from 14 different cultures in 12-h windows starting immediately after a 10-min light pulse or a control culture (thick green line) are shown. Color-matched arrows along the x axis mark the times of light pulses. (B) Immunoblot analysis of CREB and pSer-133-CREB levels in whole-cell lysates from cells harvested 15 min after a 10-min light (+) or dark (−) treatment at various circadian times. (C) ChIP assay for pCREB at the Per2 promoter CRE site shows both circadian and light modulation of its binding. Chromatin-immunoprecipitated DNA was quantified by qPCR and normalized to the signal from protein-A agarose beads. Average normalized signals (± SD; n = 3 qPCR runs) from unstimulated cells or from cells collected 30 min after 10-min light pulses are shown. ChIP signal from stimulated cells was significantly higher from control cells at all time points (*, P < 0.05 Student's t test). Data are representative of three independent experiments.
Light-activated pCREB levels did not exhibit any circadian modulation (Fig. 3B and SI Fig. 7), however, suggesting that, in our fibroblasts, gating may occur downstream of CREB activation. ChIP assays for pCREB after light stimulation at 4-h intervals showed that light at CT15 triggered a dramatic rise in binding of pCREB to the Per2 promoter CRE site (Fig. 3C). The magnitude of light-induced pCREB binding declined until CT23-CT27 and then recovered as the cycle progressed toward CT11 (or CT35). This correlated with the observed circadian gating of PER2::LUC photoinduction (Fig. 3A).
Type 0 Phase Response Curve and Singularity Behavior in Per2Luc;mOpn4 Fibroblasts.
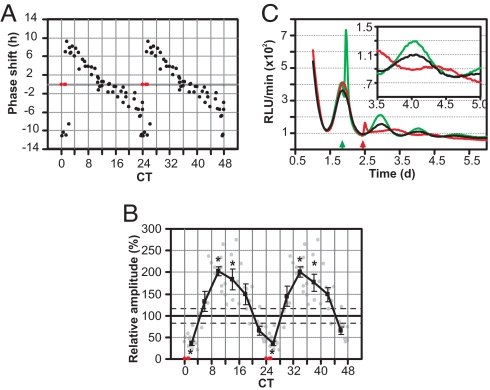
Light perturbation of Per2Luc;mOpn4 cells produced a phase-dependent resetting of PER2::LUC rhythms (SI Fig. 8). The magnitude of the phase shift plotted against the time (phase) of light pulse produced a “type 0” phase-response curve (PRC) characterized by large magnitude delays and advances (Fig. 4A).
Fig. 4.
Circadian modulation of phase shifts and amplitude changes in Per2Luc;mOpn4 fibroblasts. (A) Double-plotted PRC from 56 different light-treated samples. Each point represents the magnitude of phase shift (y axis) upon light treatment at a given time (x axis). (B) The average relative amplitude (filled square, ± SEM; n = 5–11) of the same 56 samples receiving light pulses in 4-h bins shows clear circadian modulation. Each point represents relative amplitude of individual samples (y axis) after a light pulse, expressed as percentage of amplitude of the untreated cultures during a comparable interval. Average amplitude of the untreated samples (solid horizontal line) and SEM (two parallel dashed lines, n = 5) are also shown. Statistically significant changes in amplitude (Student's t test, P < 0.05) compared with the controls are indicated by asterisks. Samples where no reliable rhythm or phase could be determined are shown in red (A and B). (C) Normalized bioluminescence traces from three representative plates that received a 10-min light pulse near CT12 (green) or CT24 (red) or received no light (black), showing a phase-dependent effect on transient induction of Per2, phase shift, and amplitude after the light pulse. For clarity, data from 3.5 to 5 days is magnified in the Inset.
Remarkably, the magnitude of the phase shift was inversely correlated with the amplitude of the PER2::LUC rhythm in the days after photostimulation (Fig. 4 B and C). A light pulse at CT12 produced no significant phase shift but did lead to a significant increase in the amplitude of the PER2::LUC rhythm. A few cultures light-pulsed at approximately CT0 showed a severe reduction in amplitude, which in some cases, reached the point of “singularity,” such that the population oscillator appeared to stop.
Phase Shift and Synchronization of Individual Cellular Oscillators.
To test whether amplitude modulation results from change in synchrony or amplitudes of individual oscillators, we monitored single cell bioluminescence from Per2Luc;mOpn4 fibroblasts (SI Fig. 9). After >30 h of bioluminescence monitoring, the cells were light-stimulated for 10 min at either the peak (CT12) or trough (CT0) of the overall luminescence rhythm (Fig. 5A–E and SI Table 1).
Fig. 5.
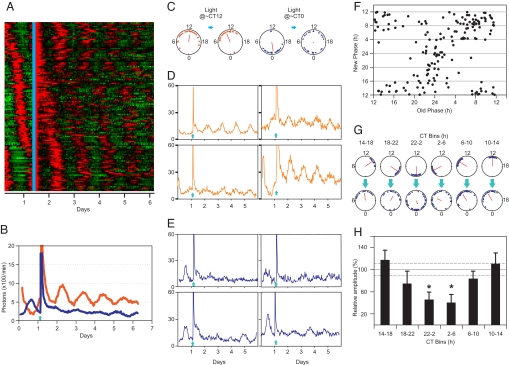
Phase shifts and amplitude changes in individual oscillators. (A) Bioluminescence heatmap of 166 fibroblasts from five different experiments in which cells received light stimulation (cyan line). (B) Overall bioluminescence from two separate cultures that received light (arrow) at the peak (orange) or the trough (blue) of the PER2::LUC rhythm show changes of amplitude in the subsequent days. (C) The respective color-coded phases (triangles) of individual fibroblasts before (Left) and after (Right) light stimulation are shown. (D and E) Sample traces of four individual cells that received light (arrow) either in the CT8–16 interval (D) or in the CT20–4 interval (E). y axis, photons/min. (F) Phase transition plot shows the phases of 166 cells (filled circles) before and after a light pulse. Light pulse at approximately CT12 resulted in small and relatively homogeneous phase shifts, whereas light at CT24 produced extremely heterogeneous phase shifts. (G) Based on the time of light pulse, cells in F were binned into six different 4-h bins. The old (Upper) and new phases (Lower) of these six groups of cells are shown in phase plots. (H) Relative average amplitudes (± SEM; n = 16–43) expressed as percentage of amplitude before light stimulation (y axis) for each of the six 4-h binned groups show circadian modulation (*, P < 0.05; Student's t test). Average amplitude before light pulse (solid horizontal line) and SEM (two parallel dashed lines, n = 166) are shown. In C and G, circadian times from 0 to 24 are plotted around the circumference of the phase plots. Arrow direction indicates average phase of the cells, and its length indicates phase coherence among cells. Values are in SI Tables 1 and 2.
The phases after light pulse plotted against the initial phases (Fig. 5F) revealed that light tends to reset the single-cell phases close to CT12. Accordingly, cells that received a light pulse near CT12 showed smaller phase shifts, whereas, with increasing phase distance from CT12, they displayed progressively larger phase shifts to achieve reset to CT12. However, some cells, particularly those near CT0 (or CT24), did not shift completely to CT12, contributing to phase desynchrony.
The extent of postpulse phase coherence depended on the time of light pulse. Based on single-cell phases at the time of light stimulation, cells were binned into six groups of 4-h bin width. The six sets of phase plots (Fig. 5G) illustrate the phase of the cells before (Upper) and after (Lower) light stimulus. In each plot, the direction and length of the mean vector represents the mean phase (μ) and coherence of the cells (r) respectively. In general, the coherence of cells after light pulse (rpostpulse) at CT10–18 was relatively high. The final coherence decreased until it reached a minimum for the CT22–2 group of cells and then increased for later phase groups (Fig. 5G and SI Table 2).
In a typical culture, the phases of individual cells at the time of photostimulation were spread over an ≈12-h interval (Fig. 5C and SI Table 1). When light was presented at CT12, the resultant single-cell phases became more coherent (rprepulse = 0.62, rpostpulse = 0.77, see SI Table 1). Conversely, light stimulation at the critical phase (CT0) reduced coherence (rprepulse = 0.76, rpostpulse = 0.05). Therefore, the oscillator synchrony is a function of both the median phase and the spread of population phase at the time of light pulse.
Amplitude Changes of Individual Cellular Oscillators.
We also measured the amplitude of single-cell bioluminescence rhythms before and after a light pulse. Average relative amplitudes for the six 4-h binned groups of cells (described above) exhibited circadian modulation (Fig. 5H). Cells receiving light stimulation at approximately CT12 did not exhibit any significant improvement in amplitude (Fig. 5D). However, cells that received light stimulation at the trough level of PER2::LUC (CT0) showed significant amplitude reduction (Fig. 5E) by as much as 40%, which, along with phase desynchronization, can further dampen the aggregate amplitude to the point of singularity.
Discussion
Fibroblasts as a Useful System for Studying Circadian Clock Resetting.
In this study, we have generated a directly light-entrainable, cell-based circadian system (Per2Luc;mOpn4 fibroblasts) that allows precise remote perturbation and longitudinal monitoring of circadian oscillators in individual cells. The Per2Luc cells are ideally suited for measuring both phase and amplitude of unit oscillators because Per2Luc bioluminescence (i) reports functionally active PER2 protein levels under transcriptional control of the native promoter (2) and (ii) exhibits more stable amplitude compared with a transiently transfected luciferase reporter (26). Also, clonal derivation of the cell line ensures uniform copy number of the mOpn4 gene in each cell. Thus, Per2Luc;mOpn4 cells qualitatively replicate key features of light signaling to the SCN, including dose-dependence of phase shifts, circadian gating of acute Per gene induction, relevant chromatin changes in the Per promoter, and suppression of overt rhythm amplitude to the point of singularity.
Gating of Acute Induction of Per2 Correlates with pCREB Recruitment to Its Promoter.
A conserved feature of most circadian oscillators is the circadian modulation of light responses. During the subjective day, photostimulation does not result in significant pCREB accumulation (7, 27, 29) or Per gene induction in the SCN. Therefore, daytime attenuation of light input may operate upstream of CREB phosphorylation, at the level of glutamate receptor signaling (30–33). However, during the mid subjective night, light triggers CREB phosphorylation but fails to induce Per transcription, implying an as yet to be determined inhibitory mechanism downstream of CREB phosphorylation.
In Per2Luc;mOpn4 fibroblasts, light-induced CREB phosphorylation does not appear to be attenuated during subjective day (Fig. 3B), consistent with the absence of the receptor-specific gating mechanism described in the SCN (30), which one would not expect in our fibroblast system. However, despite constant levels of CREB phosphorylation throughout the circadian cycle, these fibroblasts exhibit reduced induction of PER2::LUC at its circadian trough in mid night (Fig. 3A), which parallels a similar mid-night attenuation in the SCN (11), and suggests that, in both cases, a mechanism downstream of CREB phosphorylation may operate. Our findings show that reduced levels of pCREB binding at the Per2 promoter CRE site (Fig. 3C) correlate with reduced Per2 induction and may constitute a mechanism that, if operational in the SCN, can account for mid-night attenuation of Per induction.
Several questions still remain about the mechanism of circadian gating of pCREB recruitment to the Per promoter. Because the CRE site and the BMAL1/CLOCK-targeted E-box of the Per promoter are separated only by a few hundred base pairs (34), repressors bound to the E-box may sterically hinder pCREB recruitment to the CRE site. Alternatively, circadian regulation of a key CREB-interacting factor may impose circadian rhythmicity in pCREB recruitment to the CRE site.
Reciprocal Relation Between Circadian Phase Shifts and Amplitude Changes.
In mammals, photostimuli generally produce type 1 PRCs characterized by smaller phase shifts (35, 36) of the overt behavioral rhythms. Type 0 PRCs produced by brief, light stimuli are commonly described in bacteria, fungi, and plants, which harbor cell autonomous photopigments. The type 0 PRC found in this study (Fig. 4A) and others (37) favor a model in which direct activation of a cell autonomous signaling pathway can produce large phase delays and advances in individual mammalian cells, but type 1 PRCs for behavioral phase shifts result from complex intercellular interactions within the SCN.
Light stimuli also affect the amplitude of overt rhythms, which is best illustrated by the singularity phenomenon. This study revealed that an inverse relationship between phase shifts and overall amplitude changes is not restricted to the critical phase (Fig. 4). Rather, these two responses are reciprocally related throughout the circadian cycle.
Synchronization Among Individual Oscillators Contributes to Overall Amplitude.
To understand the relationships among Per gene induction, phase shifts, and amplitude changes, we monitored the effects of light stimulation on PER2::LUC rhythms in individual cells. For technical reasons (i.e., masking of bioluminescence by the light pulse itself and by phosphorescence induced by the light pulse), we were unable to estimate reliably the magnitude of acute PER2::LUC induction in each cell. Nonetheless, it seems safe to assume that the circadian gating of PER2::LUC induction as shown in Fig. 3A can only be explained by a similar gating phenomenon at the single-cell level.
Near the peak of the overall PER2::LUC rhythm, strong Per induction may bring the PER protein in individual cells to saturating and uniform levels characteristic of CT12. This would give rise to phase synchronization and a robust overall rhythm. Accordingly, daily application of brief light stimuli at CT12 in Per2Luc;mOpn4 fibroblasts produced robust PER2::LUC oscillation (SI Fig. 10). Light stimulation at other phases may cause submaximal Per induction, which could cause small or large phase shifts toward CT12. Stochastic variation in (i) the prestimulus phases of individual oscillators (Fig. 5C), and (ii) the magnitude of Per induction and phase shifts (Fig. 5 F and G) can thus generate a greater diversity of poststimulus phases. Consequently, progressively larger phase shifts are associated with reduced aggregate amplitude as one approaches CT0. At this phase of the population rhythm, individual cells are almost equally distributed across the phase-advance and phase-delay portion of the PRC (Fig. 5C). Therefore, light causes some cells to phase advance and others to phase delay with varying magnitudes. Additionally, a few cells do not show any net phase shift (Fig. 5 C, F and G). Hence, light at CT0 can disperse the phases of the individual oscillators over a large interval, such that the aggregate amplitude is severely reduced.
Amplitude Changes in Individual Oscillators.
A light pulse between CT10 and CT18 produced no significant change in amplitude, whereas the same pulse between CT22 and CT6 damped amplitude by as much as 40% (Fig. 5H). Given the phase-dependent modulation of amplitude, we cannot rule out the possibility that light could have driven a few cells to the point of singularity, but these events may be too rare to have been observed or to have made a significant contribution to the population-level singularity in our experiments. However, it raises the possibility that, in certain cell types and/or experimental conditions, single oscillator amplitude may drop to the point where no reliable rhythm can be detected. Also, it is likely that the amplitude decreases in individual cells contributed to phase dispersion: as the oscillators pass near the singularity point they can acquire diverse phases and, hence, become highly desynchronized.
A possible molecular explanation for the observed drop in amplitude may lie in a change in balance between synthesis and degradation rates of rhythmic oscillator components, or between activators and repressors of the negative feedback loop. How this altered state can be achieved and sustained over an extended time is a matter for speculation. Yet, establishing such a state in a cell culture oscillator system now offers an entry point to understand the underlying molecular mechanisms.
Phase Shifts, Amplitude Changes, and Their Adaptive Significance.
Does the inverse relationship between phase shifts and amplitude changes in fibroblast cultures apply to the coupled oscillator network in the SCN and, consequently, to whole-organism behavior? And what is its potential adaptive significance? Daily exposure to a light/dark cycle synchronizes SCN oscillators and enhances the overall amplitude of rhythmic Per gene expression (18). Additionally, rhythmic expression of the neuropeptide VIP in phase with Per genes in the SCN reinforces the molecular oscillator (38). Hence, these periodic external and internal phase-adjusting stimuli help synchronize oscillators and maintain robust rhythms. On the other hand, conditions that dampen the behavioral rhythms, such as abrupt shifts in the light/dark cycle or exposure to constant light, have been shown to desynchronize SCN oscillators (39–41). Such interplay between entraining cues and phase distribution of SCN oscillators may also form the basis for encoding photoperiod information (42, 43). Thus, phase-adjusting stimuli seem to affect the overall synchrony among coupled SCN neurons or among cultured fibroblasts in a qualitatively similar manner.
Single-cell amplitude reduction by photic stimuli at the critical circadian phase has not been directly demonstrated before in mammals. However, the clockδ19 mutation reduces oscillator amplitude and enhances sensitivity to phase shifting cues (44). Accordingly, in many organisms, the loss of overt rhythms by a critical-phase light pulse can be reversed even by weak external phase-shifting stimuli (45). Additionally, in the SCN oscillator network, robustness of damped oscillators can be restored by a few robustly rhythmic oscillators (24), which may explain the relative difficulty in inducing a prolonged singularity state in mammals. In summary, light-induced phase dispersion coupled with amplitude dampening in response to critical stimuli may offer an adaptive advantage to the animal by sensitizing it to subsequent environmental cues, so that robust rhythms may resume promptly. This phenomenon may offer an approach for therapeutic intervention in human circadian disorders.
Materials and Methods
Cell Lines and Immunoblot Analysis.
Husbandry of Per2Luc mice and fibroblast extraction from these mice were carried out following approved institutional animal care guidelines of the Salk Institute. Per2Luc;mOpn4 cells were generated by lentiviral transduction of mOpn4 into Per2Luc fibroblasts and clonal selection. Culture, maintenance, and immunoblot analysis of these cells are described in (SI Methods).
Quantitative PCR.
Total RNA from dark-control and light-exposed samples was reverse-transcribed to make cDNA (SuperScript First-Strand Synthesis System; Invitrogen), which was then subjected to SYBR green qPCR (Applied Biosystems) in a STRATAGENE MX3005P light-cycler with the primers indicated in SI Methods. Relative mRNA abundance was calculated by using the comparative δ-Ct method with β-actin mRNA as control.
Real-Time Bioluminescence Monitoring Assay.
Cells were synchronized by serum shock (23), and real-time luminescence counts from each plate were collected in a luminometer (Lumicycle; Actimetrics) as described (26). After at least 24 h of luminescence monitoring, the plates were taken out of the luminometer, placed on a different shelf of the same incubator, and exposed to 480-nm light (10-nm half peak width, 3 × 1015 photons cm−2 s−1 at plate surface) delivered via a fiber optic cable from an external 150 W halogen light source. Subsequently, the plates were returned to the luminometer for continuous bioluminescence monitoring for at least 72 h. Rhythm parameters, such as period length, phase, and amplitude were determined as described in ref. 26. Phases were calculated by using the convention that the fitted peak of the luminescence rhythm was CT12 in a 24-h cycle. All other time information, such as time of light pulse and phase shifts, were transformed to CT time.
Chromatin Immunoprecipitation.
Approximately 5 × 106 cells seeded in 150-mm cell-culture plates were synchronized with serum shock and cultured under the same conditions as those used for luminescence monitoring. Cells in 35-mm dishes were processed in parallel and monitored in the luminometer as time references. Dark-control or light-treated cells were cross-linked for 15 min with 1% formaldehyde in the dark. Subsequent steps were carried out in light as described in SI Methods.
Single-Cell Bioluminescence Imaging.
Single-cell bioluminescence imaging, image processing, and data analysis were performed as described (26). When bioluminescence rhythms from the entire field were at either peak or nadir, we remotely delivered 10-min light pulses via an optical light guide without disturbing the culture dish or interrupting image acquisition. Data were collected for at least 4 days after the light pulse. Cells exhibiting detectable bioluminescence above background during the last day of imaging were considered to be viable and were included in further analysis. Bioluminescence signals of these cells were separately analyzed to determine the amplitude, damping, phase, and period length of oscillation (26) both before and after light stimulation. These parameters were used to compute the amplitude and phase of the cell at the time of light stimulation and after light stimulation.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Victoria Piamonte for technical help, Dr. Marc Montminy (Salk Institute) for anti CREB and anti-pSer-133 CREB antibody, J.S.T. for the Per2Lucmice, and Dr. Megumi Hatori for critical evaluation of the manuscript. This work was supported by grants from the Pew Scholars program, the Whitehall Foundation, the Dana Foundation, and National Institutes of Health Grants EY016807 (to S.P.) and K08 MH067657 (to D.K.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708877104/DC1.
References
- 1.Moore RY, Eichler VB. Brain Res. 1972;42:201–206. doi: 10.1016/0006-8993(72)90054-6. [DOI] [PubMed] [Google Scholar]
- 2.Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. Proc Natl Acad Sci USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Panda S, Hogenesch JB, Kay SA. Nature. 2002;417:329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- 4.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 5.Nayak SK, Jegla T, Panda S. Cell Mol Life Sci. 2007;64:144–154. doi: 10.1007/s00018-006-5581-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meijer JH, Schwartz WJ. J Biol Rhythms. 2003;18:235–249. doi: 10.1177/0748730403018003006. [DOI] [PubMed] [Google Scholar]
- 7.Gau D, Lemberger T, von Gall C, Kretz O, Le Minh N, Gass P, Schmid W, Schibler U, Korf HW, Schutz G. Neuron. 2002;34:245–253. doi: 10.1016/s0896-6273(02)00656-6. [DOI] [PubMed] [Google Scholar]
- 8.Akiyama M, Kouzu Y, Takahashi S, Wakamatsu H, Moriya T, Maetani M, Watanabe S, Tei H, Sakaki Y, Shibata S. J Neurosci. 1999;19:1115–1121. doi: 10.1523/JNEUROSCI.19-03-01115.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tischkau SA, Mitchell JW, Tyan SH, Buchanan GF, Gillette MU. J Biol Chem. 2003;278:718–723. doi: 10.1074/jbc.M209241200. [DOI] [PubMed] [Google Scholar]
- 10.Albrecht U, Sun ZS, Eichele G, Lee CC. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 11.Yan L, Silver R. Eur J Neurosci. 2002;16:1531–1540. doi: 10.1046/j.1460-9568.2002.02224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meijer JH, Watanabe K, Schaap J, Albus H, Detari L. J Neurosci. 1998;18:9078–9087. doi: 10.1523/JNEUROSCI.18-21-09078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winfree AT. J Theor Biol. 1970;28:327–374. doi: 10.1016/0022-5193(70)90075-5. [DOI] [PubMed] [Google Scholar]
- 14.Jewett ME, Kronauer RE, Czeisler CA. Nature. 1991;350:59–62. doi: 10.1038/350059a0. [DOI] [PubMed] [Google Scholar]
- 15.Johnson CH, Kondo T. J Biol Rhythms. 1992;7:313–327. doi: 10.1177/074873049200700405. [DOI] [PubMed] [Google Scholar]
- 16.Honma S, Honma K. Am J Physiol. 1999;276:R1390–R1396. doi: 10.1152/ajpregu.1999.276.5.R1390. [DOI] [PubMed] [Google Scholar]
- 17.Covington MF, Panda S, Liu XL, Strayer CA, Wagner DR, Kay SA. Plant Cell. 2001;13:1305–1315. doi: 10.1105/tpc.13.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Challet E, Poirel VJ, Malan A, Pevet P. J Neurosci Res. 2003;72:629–637. doi: 10.1002/jnr.10616. [DOI] [PubMed] [Google Scholar]
- 19.Spoelstra K, Albrecht U, van der Horst GT, Brauer V, Daan S. J Biol Rhythms. 2004;19:518–529. doi: 10.1177/0748730404268122. [DOI] [PubMed] [Google Scholar]
- 20.Rohling J, Wolters L, Meijer JH. J Biol Rhythms. 2006;21:301–313. doi: 10.1177/0748730406290317. [DOI] [PubMed] [Google Scholar]
- 21.Schaap J, Albus H, VanderLeest HT, Eilers PH, Detari L, Meijer JH. Proc Natl Acad Sci USA. 2003;100:15994–15999. doi: 10.1073/pnas.2436298100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winfree AT. Nature. 1975;253:315–319. doi: 10.1038/253315a0. [DOI] [PubMed] [Google Scholar]
- 23.Balsalobre A, Damiola F, Schibler U. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 24.Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, et al. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirota T, Okano T, Kokame K, Shirotani-Ikejima H, Miyata T, Fukada Y. J Biol Chem. 2002;277:44244–44251. doi: 10.1074/jbc.M206233200. [DOI] [PubMed] [Google Scholar]
- 26.Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. Curr Biol. 2004;14:2289–2295. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ginty DD, Kornhauser JM, Thompson MA, Bading H, Mayo KE, Takahashi JS, Greenberg ME. Science. 1993;260:238–241. doi: 10.1126/science.8097062. [DOI] [PubMed] [Google Scholar]
- 28.Jenuwein T, Allis CD. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 29.Obrietan K, Impey S, Smith D, Athos J, Storm DR. J Biol Chem. 1999;274:17748–17756. doi: 10.1074/jbc.274.25.17748. [DOI] [PubMed] [Google Scholar]
- 30.Colwell CS. Eur J Neurosci. 2001;13:1420–1428. doi: 10.1046/j.0953-816x.2001.01517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pennartz CM, Hamstra R, Geurtsen AM. J Physiol. 2001;532:181–194. doi: 10.1111/j.1469-7793.2001.0181g.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng HY, Dziema H, Papp J, Mathur DP, Koletar M, Ralph MR, Penninger JM, Obrietan K. J Neurosci. 2006;26:12984–12995. doi: 10.1523/JNEUROSCI.4253-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng HY, Obrietan K, Cain SW, Lee BY, Agostino PV, Joza NA, Harrington ME, Ralph MR, Penninger JM. Neuron. 2004;43:715–728. doi: 10.1016/j.neuron.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 34.Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P. Proc Natl Acad Sci USA. 2002;99:7728–7733. doi: 10.1073/pnas.102075599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi JS, DeCoursey PJ, Bauman L, Menaker M. Nature. 1984;308:186–188. doi: 10.1038/308186a0. [DOI] [PubMed] [Google Scholar]
- 36.Comas M, Beersma DG, Spoelstra K, Daan S. J Biol Rhythms. 2006;21:362–372. doi: 10.1177/0748730406292446. [DOI] [PubMed] [Google Scholar]
- 37.Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 38.Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. Nat Neurosci. 2005;8:476–483. doi: 10.1038/nn1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagano M, Adachi A, Nakahama K, Nakamura T, Tamada M, Meyer-Bernstein E, Sehgal A, Shigeyoshi Y. J Neurosci. 2003;23:6141–6151. doi: 10.1523/JNEUROSCI.23-14-06141.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohta H, Yamazaki S, McMahon DG. Nat Neurosci. 2005;8:267–269. doi: 10.1038/nn1395. [DOI] [PubMed] [Google Scholar]
- 41.Albus H, Vansteensel MJ, Michel S, Block GD, Meijer JH. Curr Biol. 2005;15:886–893. doi: 10.1016/j.cub.2005.03.051. [DOI] [PubMed] [Google Scholar]
- 42.Inagaki N, Honma S, Ono D, Tanahashi Y, Honma K. Proc Natl Acad Sci USA. 2007;104:7664–7669. doi: 10.1073/pnas.0607713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.VanderLeest HT, Houben T, Michel S, Deboer T, Albus H, Vansteensel MJ, Block GD, Meijer JH. Curr Biol. 2007;17:468–473. doi: 10.1016/j.cub.2007.01.048. [DOI] [PubMed] [Google Scholar]
- 44.Vitaterna MH, Ko CH, Chang AM, Buhr ED, Fruechte EM, Schook A, Antoch MP, Turek FW, Takahashi JS. Proc Natl Acad Sci USA. 2006;103:9327–9332. doi: 10.1073/pnas.0603601103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang G, Wang L, Liu Y. EMBO J. 2006;25:5349–5357. doi: 10.1038/sj.emboj.7601397. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.