Abstract
The Caenorhabditis elegans vulva has been a valuable paradigm for defining components of signaling pathways and elucidating how signaling events are coordinated to generate a developmental pattern. Vulval precursor cells (VPCs) are induced to adopt vulval fates in the third larval stage by LIN-3, an EGF-like signal produced by the gonad. Competence to respond to the inductive signal requires that the VPCs do not fuse to the major hypodermal syncytium, hyp7. We found that two Wnt-encoding genes, cwn-1 and egl-20, play a major role in preventing fusion of VPCs with hyp7 in the second larval stage. By using tissue-specific rescue of mig-14/Wntless, which is required for the production of Wnt ligands, we found that Wnt signal produced by multiple tissues, including neurons and muscles, promotes or maintains VPC competence before vulval induction. In addition, through laser ablation and genetic analysis, we provide evidence that LIN-3 signal from the gonad also plays a significant role in preventing VPCs from fusing with hyp7. We propose that Wnt signaling plays a permissive role in preventing VPCs from fusing with hyp7 and reevaluate the roles of Wnt and LIN-3/EGF signaling in competence and induction.
Six ventral hypodermal cells named P3.p-P8.p are called vulval precursor cells (VPCs) because each is competent to respond to patterning signals that induce vulval development in the L3 stage. Previous work has suggested that competence is primarily achieved by preventing VPCs from fusing with the major hypodermal syncytium hyp7 (1–3). There appear to be points in the L1 and L2 stages when VPCs decide whether to fuse with hyp7 and thereby lose competence, or to remain independent epithelial cells and thereby maintain competence (Fig. 1).
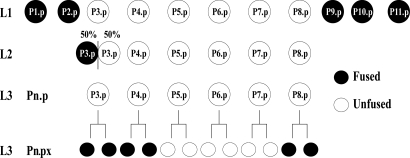
Fig. 1.
Fusion events affecting VPC competence and patterning. Pn.p cells are born during the mid-L1 stage; the anterior Pn.p cells (P1.p and P2.p) and the posterior Pn.p cells (P9.p, P10.p, and P11.p) fuse to hyp7 by the late L1 stage. In 50% of hermaphrodites, P3.p fuses to hyp7 during the L2 stage; in the other 50%, P3.p, along with P4.p and P8.p, divide after vulval induction in the L3 stage. Their daughters (“Pn.px”) fuse to hyp7 soon after birth, whereas P5.px, P6.px, and P7.px remain unfused and undergo further rounds of cell division, generating vulval cells.
The “Pn.p cells” are born in the L1 stage, and the VPCs (P3.p-P8.p) are set apart from the other ventral hypodermal cells when the anterior (P1.p and P2.p) and posterior (P9.p, P10.p, and P11.p) cells fuse with hyp7. This fusion event appears to be regulated by the activity of lin-39, which encodes a Hox protein. LIN-39 blocks fusion by repressing transcription of the fusogen eff-1 (4). The VPCs are the only Pn.p cells that express LIN-39/Hox during the L1 stage (1). Loss of lin-39 activity leads to derepression of eff-1 in P3.p-P8.p, promoting their fusion to hyp7 in the L1 stage and abrogating their competence as VPCs (4). Transcription factors that affect the restriction of lin-39 expression to P3.p-P8.p have been identified (4–6), but there is no evidence that intercellular signaling regulates lin-39 activity during the L1 stage.
In contrast, intercellular signaling maintains lin-39 activity and prevents VPCs from fusing with hyp7 during the L2 stage. In mutants lacking bar-1/β-catenin, a positive effector of the canonical Wnt pathway, some VPCs fuse inappropriately with hyp7 in the L2 stage, and LIN-39 expression is reduced (7). Forced expression of LIN-39 can overcome the bar-1 null fusion defect (7), indicating that a Wnt signaling pathway prevents VPC fusion with hyp7 through maintenance of lin-39 expression. However, in bar-1 null mutants, P5.p, P6.p, and P7.p often remain unfused with hyp7, raising the possibility that another β-catenin gene or another signaling pathway may also promote lin-39 activity and VPC competence.
Vulval induction occurs in the L3 stage when LIN-3, an EGF-like signal produced by the anchor cell of the somatic gonad, activates a canonical EGF receptor (EGFR)–Ras–MAPK cascade in P6.p, and, to a lesser extent, in P5.p and P7.p (8). Activation of the Ras cascade in P6.p leads to production of a lateral signal that activates LIN-12/Notch in P5.p and P7.p. The inductive and lateral signals pattern P5.p, P6.p, and P7.p to produce the correct numbers and types of vulval cells. P3.p, P4.p, and P8.p do not receive these patterning signals and produce daughters that fuse with hyp7. lin-39 also appears to be involved in induction and is a direct transcriptional target of the inductive signaling pathway (9); it also appears to be involved in execution of vulval fates (1, 2, 7). The multiple requirements for lin-39 in vulval development complicate the interpretation of mutant phenotypes of genes involved in regulating lin-39, and hence in distinguishing between their potential roles in competence as opposed to induction of vulval fates.
Here, we focused specifically on the roles of Wnt and EGF signaling in competence by scoring VPC fusion with hyp7 before induction. We find that cwn-1 and egl-20 are the major Wnt ligands that regulate this process, and that multiple tissues redundantly contribute Wnt signal to promote competence. Finally, we provide evidence that, in addition to initiating vulval development, the EGF-like inductive signal LIN-3 also promotes VPC competence. We also discuss the implications of our results for the roles of Wnt and LIN-3/EGF signaling in promoting competence and induction.
Results
The main criterion we use to assess competence is whether a VPC has fused with hyp7 or not. Here, we use the term “F fate” (F for “fused”) to indicate that a VPC had fused with hyp7 in the L2 stage, as in its original usage (7). The term “3° fate” denotes a VPC that divides in the L3 stage to produce two daughters that fuse with hyp7, and “vulval fate” or “induced” denotes a VPC that undergoes more than one round of cell division and produces descendants that do not fuse with hyp7 (8). In all experiments, we use dpy-7p::2nls::yfp [transgenes arIs99 and arIs100] to assess cell fusion; these transgenes mark all hyp7 nuclei, including Pn.p and Pn.px nuclei that have become part of hyp7 (10). dpy-7p::2nls::yfp expression correlates with loss of the adherens junction marker AJM-1::GFP (11) (T.R.M., M.-S. Choi, and I.G., unpublished observations), confirming its efficacy as a fusion marker.
Compromised Wnt Gene Activity Causes VPCs to Fuse with hyp7 in the L2 Stage.
Mutations in bar-1/β-catenin implicated Wnt signaling in promoting competence (7). To identify the relevant Wnt ligand(s), we surveyed the five predicted Wnt-encoding genes in Caenorhabditis elegans: cwn-1/Wnt4, cwn-2/Wnt5B, egl-20/Wnt16, mom-2/Wnt5A, and lin-44/Wnt7B (12). We analyzed predicted null mutations for cwn-1, cwn-2, and lin-44 at 20°C and severe temperature-sensitive loss-of-function mutations for egl-20 and mom-2 at 25°C, the restrictive temperature (13). We found that some VPCs in cwn-1(ok546), egl-20(n585), and cwn-2(ok895) single mutants inappropriately fuse with hyp7 in the L2 stage (Table 1) (14). Although P3.p and P4.p are both affected in these single mutants, the phenotype is most evident for P4.p, which never fuses with hyp7 in wild type, but fuses 87% of the time in cwn-1, 55% in egl-20 (grown at the restrictive temperature, 25°C) and 6% in cwn-2 (Table 1). The frequency of P4.p fusion is 100% in the cwn-1;egl-20 double mutant grown at 25°C (Table 1); in this strain, P5.p, P7.p, and P8.p also show a significant frequency of inappropriate fusion with hyp7. These results suggest that cwn-1 and egl-20 are the main Wnt ligands involved in preventing inappropriate fusion of P4.p with hyp7, thereby maintaining VPC competence into the L3 stage.
Table 1.
Wnt signaling maintains VPC competence in the L2 stage
| Mutants | Fused/tertiary/induced,* % |
N | |||||
|---|---|---|---|---|---|---|---|
| P3.p | P4.p | P5.p | P6.p | P7.p | P8.p | ||
| arIs99† | 50/50/0 | 0/100/0 | 0/0/100 | 0/0/100 | 0/0/100 | 0/100/0 | 60 |
| arIs100† | 52/48/0 | 0/100/0 | 0/0/100 | 0/0/100 | 0/0/100 | 0/100/0 | 42 |
| cwn-1(ok546) | 98/2/0 | 87/11/0 | 11/0/89 | 0/0/100 | 0/8/92 | 2/98/0 | 110 |
| egl-20(n585ts)‡ | 96/4/0 | 54/45/0 | 3/0/97 | 0/0/100 | 0/3/97 | 1/99/0 | 75 |
| cwn-2(ok895) | 77/23/0 | 6/93/0 | 2/0/98 | 0/0/100 | 0/1/99 | 2/98/0 | 111 |
| mom-2(ne874ts)§ | 51/49/0 | 0/100/0 | 0/0/100 | 0/0/100 | 0/0/100 | 0/100/0 | 103 |
| lin-44(n1792) | 39/61/0 | 1/99/0 | 0/0/100 | 0/0/100 | 0/0/100 | 0/100/0 | 113 |
| cwn-1(ok546); egl-20(n585ts)‡ | 96/0/0 | 94/0/1 | 26/0/73 | 4/0/90 | 16/11/57 | 11/56/0 | 73 |
| mig-14(ga62) | 100/0/0 | 96/4/0 | 21/3/76 | 3/1/96 | 6/24/70 | 12/88/0 | 101 |
Wnt signaling appears to compromise the L2 fusion event: we staged mig-14(ga62) hermaphrodites by using a marker that is expressed during molting events (mlt-10::gfp)(40), and found that VPCs fuse either before or during the L2 molt (data not shown). For routine scoring of different genotypes, shown here, L3 hermaphrodites were scored for expression of GFP in P3.px-P8.px or their descendants so that the 3° or induced fates could also be scored. However, spot-checks of L1 and L2 stage worms of all genotypes indicate that fusion does not occur in the L1 stage and can occur in the L2 stage.
*The percentage occasionally does not equal 100%, because cells were missing or had other abnormalities.
†All wnt mutants, except for mom-2(ne874ts), carry the hypodermal marker arIs99[dpy-7::2Xnls::yfp]. mom-2(ne874ts) carries arIs100[dpy-7::2Xnls::yfp].
‡Grown at the nonpermissive temperature, 25°C.
§mom-2(ne874ts) was grown at 15°C, and bleached mixed-stage eggs were transferred to the nonpermissive temperature.
Another study (15) concluded that cwn-1, egl-20, and all of the other Wnt-encoding genes are functionally redundant for “VPC specification” after detecting synergy between cwn-1 and mutations in the four other genes. They reported that P5.p-P7.p were “underinduced” (i.e., adopted either the F or the 3° fate, as opposed to a vulval fate). However, they did not distinguish between the F and 3° fates, and therefore did not distinguish whether a VPC could have been induced or not. For example, if P5.p had adopted the F fate, it would not have been available to receive the inductive signal. Furthermore, 3° fate could reflect reduced competence to respond to the inductive signal rather than lack of induction per se (see Discussion).
Multiple Tissues Are Redundant Sources of Wnt Activity for Promoting VPC Competence.
Identifying the cellular source of the Wnt signals that prevent VPC fusion is critical for understanding the cell–cell interactions that underlie VPC fate specification. Transcriptional reporters indicate that wnt genes, including cwn-1 and egl-20, are expressed in many different tissues and at different times during development (refs. 15–19; T.R.M. and I.G., unpublished observations). However, expression patterns are not a reliable indicator of the cellular focus of action, because transcriptional reporters may not contain all sequences that direct transcription of the endogenous genes, can only be visualized if the expression levels are relatively high, and do not reveal potential posttranscriptional regulation affecting production of functional ligand.
To determine the cellular focus of Wnt signaling for preventing VPC fusion, we considered several approaches. Mosaic analysis has severe limitations in this context: for example, neurons, hyp7, and VPCs are all produced from the ABp lineage, and losses in combinations of lineages would be difficult to obtain. Rescue of cwn-1 or egl-20 single or double mutants using tissue-specific promoters might provide misleading information, because Wnt proteins are secreted ligands, and might function if produced in nearby cells, even if they do not normally serve as the source for promoting VPC competence.
Tissue-specific rescue of mutations affecting factors required for Wnt production in the source cell circumvents these potential problems, as well as the additional problem of genetic redundancy of Wnt ligands. The first such factor identified was MOM-1/Porcupine (20); however, we could not assess the focus of mom-1, because a null allele is maternal effect lethal (21), and homozygotes segregating from heterozygous mothers do not have a vulval defect (data not shown). Another factor is Wntless/Evi (mig-14 in C. elegans), which was demonstrated to be required in Wnt-producing cells for proper secretion of Wnt ligands (22, 23). mig-14(ga62), a viable partial loss-of-function allele, affects VPC fate (24), causing a highly penetrant fusion defect for P3.p and P4.p and a lower penetrance defect for some other VPCs (Table 1).
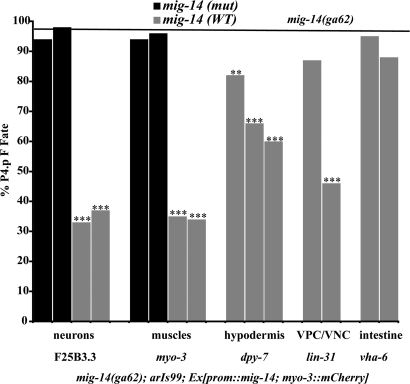
We analyzed transgenes expressing MIG-14(+) in specific tissues for their ability to rescue the inappropriate fusion of P4.p to hyp7 in a mig-14(ga62);arIs99 background (Fig. 2). We used well characterized tissue-specific promoters that are expressed continuously in muscles, neurons, hypodermis, or intestine from the L1 stage through the time of vulval induction to drive MIG-14(+) expression. At least one wnt is expressed in each of these tissues (refs. 15–19; T.R.M. and I.G., unpublished observations). Our results demonstrate that expression of MIG-14 in muscles, neurons, or hypodermis can partially rescue the P4.p defect (Fig. 2), suggesting that Wnt signaling from all of these sources contributes to VPC competence.
Fig. 2.
mig-14 acts in multiple tissues to regulate VPC fusion with hyp7. Rescue experiments were performed in a mig-14;arIs99 background. The transgenic marker used for all lines was myo-3p::mCherry. Each bar represents a single transgenic line. The black bars represent negative control lines in which a promoter drives expression of a mutated mig-14 cDNA that does not make functional protein. The gray bars represent experimental lines in which a promoter drives a wild-type mig-14 cDNA that encodes a functional protein. dpy-7p::mig-14cDNA was injected at 5 μg/ml (the first two gray bars) and 10 μg/ml (the third gray bar). Animals were scored for whether P4.p fuses directly with hyp7, and the results for each line were compared against the P4.p defect observed in mig-14;arIs99, which is represented by the horizontal black line. P values were calculated by using a z test to compare the proportion of abnormal animals in each group. ***, P < 0.001; **, P < 0.01. n ≥ 50 for all lines.
We could not assess whether the gonad might produce a Wnt ligand that promotes competence because of the lack of a somatic gonad-specific promoter. Although there are no reported cases of gonadal expression of Wnt genes during the L1 or L2 stages, conclusions based on reporter gene analysis have the caveats enumerated above. As described in the next section, we believe that the somatic gonad serves as the source of a different competence-promoting signal, LIN-3.
lin-3/EGF Signaling and the Gonad Contribute to Preventing P5.p, P6.p, and P7.p Fusion with hyp7.
In cwn-1;egl-20 hermaphrodites grown at 25°C, P6.p almost never fuses with hyp7, and P5.p and P7.p sometimes fuse, although less frequently than P4.p. Although the increased susceptibility of more anterior Pn.p cells to adopt the F fate when Wnt signaling is reduced has been attributed to a lower level of a graded Wnt signal emanating from the posterior region (15), we hypothesized that this pattern might reflect a graded signal produced by the gonad before induction. Even though vulval induction does not occur until the L3 stage, lin-3 is transcribed in the presumptive anchor cell as well as other cells in the central region of the gonad in the L2 stage (25), making it a good candidate for such a signal. Interestingly, reducing let-23 activity in a bar-1 mutant background also causes P5.p, P6.p, and P7.p to fuse more frequently (7), an observation that could be consistent with our hypothesis or with alternative possibilities of constitutive let-23 basal activity or an external signaling input other than lin-3 (26).
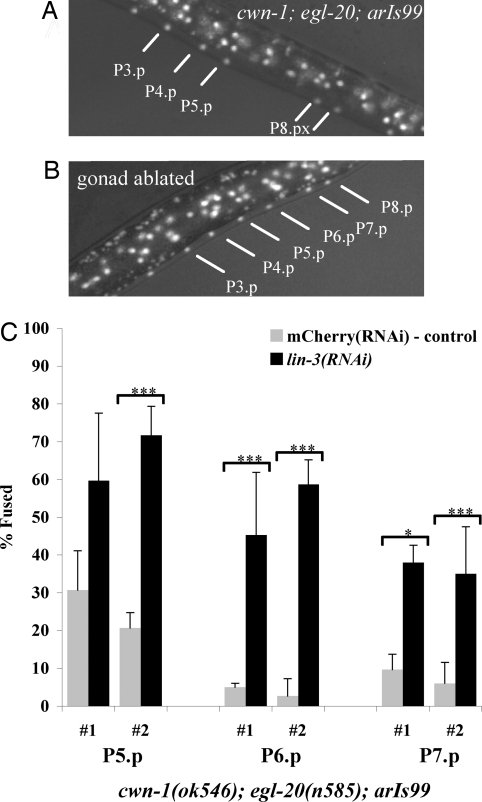
To test this hypothesis, we assessed the potential contributions of the gonad and of lin-3 activity to competence. We ablated the gonad in L1 cwn-1;egl-20;arIs99 hermaphrodites and found that P5.p, P6.p, and P7.p adopted the F fate more frequently: P5.p adopted the F fate in four of five individuals, P6.p adopted the F fate in three of five individuals, and P7.p adopted the F fate in one of five individuals, as opposed to mock-operated animals in which these VPCs did not adopt the F fate (zero of five). Furthermore, P5.p adopted the F fate when we ablated the gonads in L1 egl-20;arIs99 single mutants (three of four). Thus, ablation of the gonad promotes fusion of these VPCs with hyp7, implying a role for the gonad in promoting VPC competence. The relative resistance of P7.p to fusion may reflect the presence of mab-5/Hox activity (1).
We performed lin-3(RNAi) in cwn-1;egl-20;arIs99 and control arIs99 hermaphrodites. lin-3(RNAi) in control hermaphrodites caused VPCs to adopt the 3° fate, as expected for loss of inductive signal, but did not cause fusion to hyp7 (data not shown). lin-3(RNAi) enhanced the phenotype of the Wnt double mutant so that P5.p, P6.p, and P7.p fused to hyp7 more frequently relative to the control (Fig. 3). Reducing lin-3 activity appears to have the greatest effect on the competence of P6.p, which almost always remains unfused otherwise, consistent with higher lin-3 signaling to P6.p (see below).
Fig. 3.
The gonad and lin-3/EGF promote the competence of P5.p, P6.p, and P7.p. (A) An L3 “Pn.pxx-stage” cwn-1;egl-20;arIs99 hermaphrodite; lines point to VPC nuclei labeled with GFP, indicating that they have fused with hyp7 (here, P3.p, P4.p, and P5.p fused with hyp7; P6.p and P7.p adopted vulval fates; and P8.p adopted the 3° fate). (B) Individual cwn-1;egl-20;arIs99 hermaphrodite in which gonad ablation caused P5.p-P7.p to fuse with hyp7 (see text). (C) lin-3(RNAi) reduced the competence of P5.p-P7.p when wnt activity is compromised. The gray bars represent lin-3(RNAi), and the black bars represent mCherry(RNAi) in a cwn-1;egl-20;arIs99 background. Each set represents a separate RNAi experiment. A t test was used to determine the P values and standard deviation of the P5.p, P6.p, and P7.p cells that adopt the F fate compared with the negative control. ***, P < 0.001; **, P < 0.01; and *, P < 0.05. n > 60 for all experiments.
The results of the gonad ablation and lin-3(RNAi) experiments taken together suggest that lin-3 signaling from the gonad helps to maintain competence of P5.p, P6.p, and P7.p by preventing them from fusing with hyp7. In this context, the synergy between bar-1 and let-23 is consistent with the “inductive signaling pathway” operating in the L2 stage to promote competence. Furthermore, we find that a reporter for inductive signaling activity, arIs92[egl-17p::cfp-lacZ], is expressed in L2 hermaphrodites, strongly in P6.p and more weakly in flanking VPCs. This expression depends on the presence of the gonad: in intact hermaphrodites, 13 of 13 express CFP-LacZ in P6.p, whereas zero of four express CFP-LacZ in P6.p after the gonad has been ablated. Taken together, the results suggest that LIN-3 activity in the L2 stage activates the canonical RTK–Ras–MAPK cascade that underlies vulval induction to promote competence.
Discussion
To be competent to adopt a vulval fate, a VPC must remain unfused with hyp7, be able to progress through the cell cycle and divide, and be able to respond to the inductive signal. lin-39 activity is important for all three of these features, as well as in execution of vulval fates (reviewed in ref. 8). Another aspect of competence is the availability of the receptors for the inductive and lateral signals and other factors needed for signal transduction: these components must be transcribed, translated, modified, and localized appropriately to the apical or basolateral domain so that activating ligands elicit signal transduction. Whether lin-39 modulates this aspect of competence for vulval induction is not known, although it has been implicated in the expression of lin-12 and therefore for competence to respond to the lateral signal (27). Here, we discuss our results in terms of whether and how Wnt and LIN-3 signaling promote competence and induction. The central role of lin-39 for different aspects of competence, and its position as a target of both the Wnt and LIN-3 signaling pathways, must be borne in mind when considering the roles of LIN-3/EGF and Wnt signaling in maintaining competence and initiating induction.
Wnt Signaling and VPC Competence.
Several previous studies indicated that loss of the canonical Wnt signal transduction pathway in the VPCs causes inappropriate fusion with hyp7 (1, 2, 7, 15, 28). We have shown that the Wnt ligands encoded by cwn-1 and egl-20 are functionally semiredundant and are the major Wnts for maintaining VPC competence in the L2 stage; our findings are consistent with data for cwn-1 and egl-20 presented in another study, which also suggested that other Wnt ligands have additional, but minor roles in maintaining competence (15). In addition, we have demonstrated that several tissues provide functionally relevant sources of Wnt signaling activity to promote VPC competence. These observations contrast with many other systems in which precise spatial control of specific Wnt proteins is critical for patterning (29). We suggest that the multifocal origin and high degree of functional redundancy among Wnt signals indicates that Wnt signaling functions as a permissive or trophic factor, rather than as a patterning agent, in maintaining VPC competence.
LIN-3/EGF Signaling and VPC Competence.
We have shown that lin-3, the inductive signal that specifies VPCs to adopt vulval fates in the L3 stage, also promotes competence, as lin-3(RNAi) or gonad ablation performed in the egl-20 single mutant or cwn-1;egl-20 double mutant background caused more VPCs to fuse with hyp7 before induction. Synergy between let-23 and bar-1 (7) and gonad-dependent expression of a canonical inductive pathway target gene, egl-17, in the VPCs in the L2 stage (see Results) together suggest that the canonical RTK–Ras–MAPK cascade that underlies vulval induction also underlies competence, perhaps by promoting lin-39 expression.
At the time of vulval induction, egl-17 transcription in response to the inductive signal is strongly and consistently visualized in P6.p (30), and transiently visualized in P5.p and P7.p but then lost after lateral signaling (31). It may be that the graded expression of egl-17::gfp observed at the beginning of the L3 stage is the residue of LIN-3 activity during the L2 stage. However, continued lin-3 activity in the L3 stage may also contribute to competence at the time of vulval induction, perhaps by “taking over” the role of Wnt signaling in promoting lin-39 expression. Indeed, lin-3 may function in both the L2 and early L3 stages to promote competence. We cannot assess these possibilities at this time.
LIN-3/EGF Signaling: Maintaining Competence Versus Initiating Vulval Induction.
There is abundant evidence indicating that lin-3 induces vulval fates (8). Alterations in the activity of lin-3 or components of the EGFR–Ras–MAPK signal transduction pathway affect vulval induction: loss of activity causes VPCs that normally adopt vulval fates to instead adopt the 3° fate, whereas excess activity causes VPCs that normally adopt the 3° fate to instead adopt vulval fates. Furthermore, lin-3 is expressed in the anchor cell of the gonad, which in a wild-type background is necessary and sufficient for vulval induction in the L3 stage.
In an otherwise wild-type background, loss of lin-3 or the gonad does not cause VPCs to fuse with hyp7, indicating that the major role of lin-3 and the gonad is vulval induction rather than promoting competence. However, we have shown that, under conditions of limited wnt signaling, loss of lin-3 activity or ablation of the gonad causes VPCs to fuse with hyp7. Our observations suggest that lin-3 signal from the gonad promotes VPC competence in addition to inducing vulval development (Fig. 4).
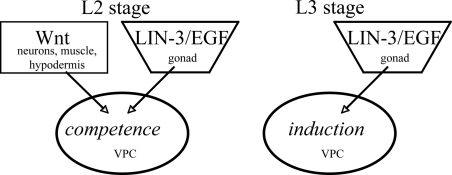
Fig. 4.
Inferred roles for Wnt and LIN-3/EGF signaling in VPC competence and induction. At this time, we believe that roles for Wnt and LIN-3 in competence and for LIN-3 in induction have been substantiated, but a role for Wnt signaling in induction remains an open question (see Discussion).
A dual role for lin-3 in promoting competence as well as induction raises the questions of whether and how competence and induction differ on the molecular level. Competence may differ from induction simply in the absolute level of lin-39 activity; however, expression of LIN-39 under the control of a heat-shock promoter is not sufficient to cause vulval induction and cannot bypass the need for the inductive signal (2). Instead, we favor the view that, in the L3 stage, increased production or secretion of LIN-3 from the anchor cell, increased sensitivity of VPCs to LIN-3, or other differences in VPCs, activates additional targets that drive or constitute induction. It is also possible that there are unique target genes of the EGFR–Ras–MAPK pathway in the L2 stage. An analysis of the contribution of different target genes of the EGFR–Ras–MAPK pathway to competence and induction should help clarify this issue.
Wnt Signaling: Maintaining Competence Versus Promoting Vulval Induction.
A major role for Wnt signaling in maintaining or promoting competence is clear: reducing Wnt signaling activity causes VPCs to fuse directly with hyp7. In addition, preliminary results indicate that posterior Pn.p cells fail to fuse with hyp7 in pry-1/Axin mutants: for example, P9.p and P10.p always fuse in a wild-type background, but fail to fuse ≈35% of the time in pry-1(mu38). These results suggest that constitutive activation of Wnt signaling generally acts as a block to Pn.p fusion with hyp7.
In analyzing mutants with reduced wnt activity, the major role of Wnt signaling in competence masks any putative later role in vulval induction. In our analysis, the failure of P5.p, P6.p, or P7.p to adopt a vulval fate generally did not reflect a failure of induction per se. Instead, in most cases, when P5.p, P6.p, and/or P7.p was not induced, it had adopted the F fate and therefore was no longer present to be induced; given the strong and documented effect of Wnt activity on competence, the cases in which P5.p, P6.p, or P7.p adopted the 3° fate may indicate a reduced ability to respond to the inductive signal because of reduced competence rather than lack of induction. The high degree of functional redundancy among Wnt genes and pathway components, their essential roles in other aspects of development, and the multifocal nature of Wnt signal production make it essentially impossible to achieve a truly null situation, further complicating inferences based on loss-of-function phenotypes.
Gain-of-function situations are also problematic. Constitutively active Wnt signal transduction causes VPCs that normally adopt the 3° fate to be “overinduced” (as assessed by ectopic invaginations), even when EGFR–Ras–MAPK activity is reduced (28). However, a situation in which Wnt signaling is artificially activated may be misleading as an indication of its normal role; a cautionary tale is afforded by the effect of removing SynMuv gene activity, which creates an artificial signaling event (32). In addition, lin-39 activity both prevents fusion and promotes division (4); thus, it is conceivable that abnormally high lin-39 levels in pry-1 mutants (perhaps in combination with other ectopically elevated factors) could cause spurious division so that P4.p or P8.p may divide ectopically without having responded to normal patterning signals.
For these reasons, it is not clear whether and how Wnt signaling contributes to vulval induction as opposed to competence. One way to distinguish between these roles would be to identify targets of Wnt signaling that differentially affect only competence or only induction. At this time, however, the only known Wnt target is lin-39, which is also a target of LIN-3/EGF. Therefore, we believe that, currently, the only well substantiated role for Wnt signaling is to maintain VPC competence (Fig. 4).
Materials and Methods
Strains and Genetic Analysis.
C. elegans var. Bristol strain N2 (33) was the wild-type parent strain of all mutants and markers used. The mutations used for analysis include the following: LGI: lin-44(n1792) (34); LGII: arIs100[dpy-7::yfp;ceh-22::gfp] (10), cwn-1(ok546), mig-14(ga62) (24); LGIV: egl-20(n585ts) (35), cwn-2(ok895); LGV: mom-2(ne874ts) (13); LGX: arIs99[dpy-7::yfp;ceh-22::gfp] (10). The recipient used for creating transgenes was GS4441 mig-14(ga62);arIs99. All strains were grown by using standard procedures at 20°C (33), except for mom-2(ne874ts), which was grown at 15°C and egl-20(n585), which was grown at 25°C. cwn-1(ok546) was backcrossed three times.
Constructs.
mig-14(+) cDNA was synthesized (DNA 2.0, Menlo Park, CA), based on the sequence given for R06B9.6 (www.wormbase.org), with 5′ and 3′ BglII sites. The cDNA was cloned into pDONR221. All constructs for tissue-specific rescue [see supporting information (SI)] were constructed in the form of tissue-specific promoter::mig-14(+) cDNA::unc-54 3′-UTR. The promoters used drive expression continuously from the L1 stage in the following tissues: dpy-7, hyp7 (10); myo-3, body wall muscle (Fire Vector kit 1999); F25B3.3, most neurons (36); vha-6, intestine (37). lin-31p drives expression in the VPCs (10), but with a more sensitive reporter, mCherry, it also appears to drive expression in the ventral nerve cord and hence is not strictly VPC-specific (N. deSouza, T.R.M., and I.G., unpublished observations).
Transgenic Lines.
The recipient strain for mig-14 tissue-specific promoter rescue experiments was GS4441 [mig-14(ga62);arIs99]. myo-3p::mCherry (20 μg/ml) was used as the coinjection marker for all transgenic lines created. All injection mixes contain pBluescript to make a total DNA concentration of 100 μg/ml. dpy-7p::mig-14 was microinjected at a concentration of 5 μg/ml. myo-3p constructs were microinjected at a concentration of 20 μg/ml. lin-31p::mig-14 was microinjected at a concentration of 30 μg/ml. F25B3.3p constructs were microinjected at a concentration of 50 μg/ml. vha-6p::mig-14 was microinjected at a concentration of 20 μg/ml. Individual F1 worms were selected for the presence of mCherry expression in body wall muscles to establish independent extrachromosomal lines.
VPC Fate Determination.
Worms were scored from L3 Pn.px to Pn.pxx stage to ensure that competent VPCs could have been induced. All strains also contained arIs99, and VPCs or Pn.px cells were considered fused when nuclei displayed YFP.
We note that there are additional morphological abnormalities in cwn-1;egl-20;arIs99 worms. Nuclei of Pn.p cells were often mislocated or occasionally missing. The use of arIs99 made it much easier to score the VPC fates given these abnormalities.
We performed experiments with egl-20(n585ts) at 25°C, the restrictive temperature. The P4.p fusion defect is greatest at this temperature as well. However, surprisingly, cwn-1;egl-20(n585);arIs99 hermaphrodites and mig-14 hermaphrodites have a more severe VPC fusion defect at 15°C than at 25°C or 20°C, respectively, suggesting that competence may be inherently cold-sensitive.
Gonad Ablations.
Cell ablations were performed on worms grown at 25°C. Newly hatched L1 larvae were placed on 2% agarose pads in M9 with 10 mM sodium azide, and gonadal nuclei were ablated with a laser microbeam. The absence of a gonad was confirmed at the time the effect was scored. For cwn-1;egl-20;arIs99, fusion defects were assayed during the L4 stage; for arIs92[egl-17p::cfp-lacZ] hermaphrodites, expression in P6.p was scored in the L2 stage.
RNAi Analysis.
cwn-1;egl-20;arIs99 worms grown at 25°C were fed bacteria expressing dsRNA (38). lin-3 dsRNA (39) or control mCherry dsRNA bacteria was applied to plates containing isopropyl β-d-thiogalactoside and ampicillin. After 5 h, several L4 worms were put onto the plates. F1 progeny were scored for fusion defects between the Pn.px and L4 stages. The experiment was performed on two separate days, and three individual plates were scored for each variable.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Cory Dunn, Chip Ferguson, Jennifer Green, Ji Li, Michael Stern, Meera Sundaram, and Paul Sternberg for discussion and comments on the manuscript; Katherine Lelli for constructing and scoring pry-1;arIs99; the Caenorhabditis Genetics Center (St. Paul, MN), the C. elegans Gene Knockout Consortium (Oklahoma City, OK, and Vancouver, Canada), and the National Bioresource Project for the Nematode C. elegans (Tokyo, Japan) for strains; Xinlan Zhou for performing microinjections; Richard Ruiz for technical assistance; and David Eisenmann for exchanging results before publication. This work was supported by National Institutes of Health Grant CA095389 (to I.G.). I.G. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709989104/DC1.
References
- 1.Clandinin TR, Katz WS, Sternberg PW. Dev Biol. 1997;182:150–161. doi: 10.1006/dbio.1996.8471. [DOI] [PubMed] [Google Scholar]
- 2.Maloof JN, Kenyon C. Development. 1998;125:181–190. doi: 10.1242/dev.125.2.181. [DOI] [PubMed] [Google Scholar]
- 3.Wang M, Sternberg PW. Dev Biol. 1999;212:12–24. doi: 10.1006/dbio.1999.9357. [DOI] [PubMed] [Google Scholar]
- 4.Shemer G, Podbilewicz B. Genes Dev. 2002;16:3136–3141. doi: 10.1101/gad.251202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alper S, Kenyon C. Development. 2001;128:1793–1804. doi: 10.1242/dev.128.10.1793. [DOI] [PubMed] [Google Scholar]
- 6.Alper S, Kenyon C. Development. 2002;129:3335–3348. doi: 10.1242/dev.129.14.3335. [DOI] [PubMed] [Google Scholar]
- 7.Eisenmann DM, Maloof JN, Simske JS, Kenyon C, Kim SK. Development. 1998;125:3667–3680. doi: 10.1242/dev.125.18.3667. [DOI] [PubMed] [Google Scholar]
- 8.Sternberg PW. WormBook. 2005 Jun 25; doi: 10.1895/wormbook.1.6.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagmaister JA, Gleason JE, Eisenmann DM. Mech Dev. 2006;123:135–150. doi: 10.1016/j.mod.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Myers TR, Greenwald I. Dev Cell. 2005;8:117–123. doi: 10.1016/j.devcel.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 11.Koppen M, Simske JS, Sims PA, Firestein BL, Hall DH, Radice AD, Rongo C, Hardin JD. Nat Cell Biol. 2001;3:983–991. doi: 10.1038/ncb1101-983. [DOI] [PubMed] [Google Scholar]
- 12.Eisenmann DM. WormBook. 2005 Jun 25; doi: 10.1895/wormbook.1.7.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakamura K, Kim S, Ishidate T, Bei Y, Pang K, Shirayama M, Trzepacz C, Brownell DR, Mello CC. Genes Dev. 2005;19:1749–1754. doi: 10.1101/gad.1323705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frand AR, Russel S, Ruvkun G. PLoS Biol. 2005;3:e312. doi: 10.1371/journal.pbio.0030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gleason JE, Szyleyko EA, Eisenmann DM. Dev Biol. 2006;298:442–457. doi: 10.1016/j.ydbio.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 16.Herman MA, Vassilieva LL, Horvitz HR, Shaw JE, Herman RK. Cell. 1995;83:101–110. doi: 10.1016/0092-8674(95)90238-4. [DOI] [PubMed] [Google Scholar]
- 17.Inoue T, Oz HS, Wiland D, Gharib S, Deshpande R, Hill RJ, Katz WS, Sternberg PW. Cell. 2004;118:795–806. doi: 10.1016/j.cell.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Pan CL, Howell JE, Clark SG, Hilliard M, Cordes S, Bargmann CI, Garriga G. Dev Cell. 2006;10:367–377. doi: 10.1016/j.devcel.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 19.Whangbo J, Kenyon C. Mol Cell. 1999;4:851–858. doi: 10.1016/s1097-2765(00)80394-9. [DOI] [PubMed] [Google Scholar]
- 20.Coudreuse D, Korswagen HC. Development. 2007;134:3–12. doi: 10.1242/dev.02699. [DOI] [PubMed] [Google Scholar]
- 21.Thorpe CJ, Schlesinger A, Carter JC, Bowerman B. Cell. 1997;90:695–705. doi: 10.1016/s0092-8674(00)80530-9. [DOI] [PubMed] [Google Scholar]
- 22.Banziger C, Soldini D, Schutt C, Zipperlen P, Hausmann G, Basler K. Cell. 2006;125:509–522. doi: 10.1016/j.cell.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 23.Bartscherer K, Pelte N, Ingelfinger D, Boutros M. Cell. 2006;125:523–533. doi: 10.1016/j.cell.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 24.Eisenmann DM, Kim SK. Genetics. 2000;156:1097–1116. doi: 10.1093/genetics/156.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill RJ, Sternberg PW. Nature. 1992;358:470–476. doi: 10.1038/358470a0. [DOI] [PubMed] [Google Scholar]
- 26.Chen Z, Han M. Curr Biol. 2001;11:1874–1879. doi: 10.1016/s0960-9822(01)00596-6. [DOI] [PubMed] [Google Scholar]
- 27.Takacs-Vellai K, Vellai T, Chen EB, Zhang Y, Guerry F, Stern MJ, Muller F. Dev Biol. 2007;302:661–669. doi: 10.1016/j.ydbio.2006.09.049. [DOI] [PubMed] [Google Scholar]
- 28.Gleason JE, Korswagen HC, Eisenmann DM. Genes Dev. 2002;16:1281–1290. doi: 10.1101/gad.981602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hecht A, Kemler R. EMBO Rep. 2000;1:24–28. doi: 10.1093/embo-reports/kvd012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burdine RD, Branda CS, Stern MJ. Development. 1998;125:1083–1093. doi: 10.1242/dev.125.6.1083. [DOI] [PubMed] [Google Scholar]
- 31.Yoo AS, Bais C, Greenwald I. Science. 2004;303:663–666. doi: 10.1126/science.1091639. [DOI] [PubMed] [Google Scholar]
- 32.Cui M, Chen J, Myers TR, Hwang BJ, Sternberg PW, Greenwald I, Han M. Dev Cell. 2006;10:667–672. doi: 10.1016/j.devcel.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 33.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herman MA, Horvitz HR. Development. 1994;120:1035–1047. doi: 10.1242/dev.120.5.1035. [DOI] [PubMed] [Google Scholar]
- 35.Trent C, Tsuing N, Horvitz HR. Genetics. 1983;104:619–647. doi: 10.1093/genetics/104.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aurelio O, Boulin T, Hobert O. Development. 2003;130:599–610. doi: 10.1242/dev.00277. [DOI] [PubMed] [Google Scholar]
- 37.Oka T, Toyomura T, Honjo K, Wada Y, Futai M. J Biol Chem. 2001;276:33079–33085. doi: 10.1074/jbc.M101652200. [DOI] [PubMed] [Google Scholar]
- 38.Timmons L, Court DL, Fire A. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 39.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.