Abstract
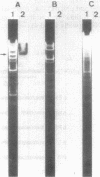
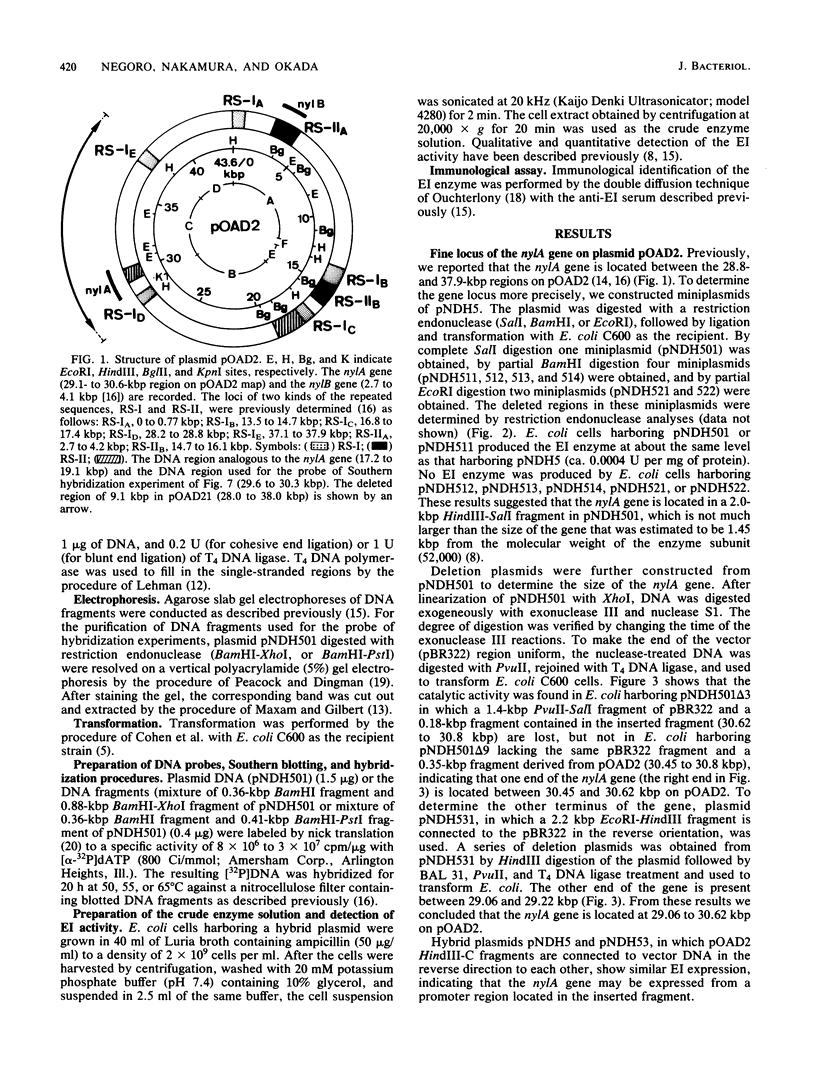
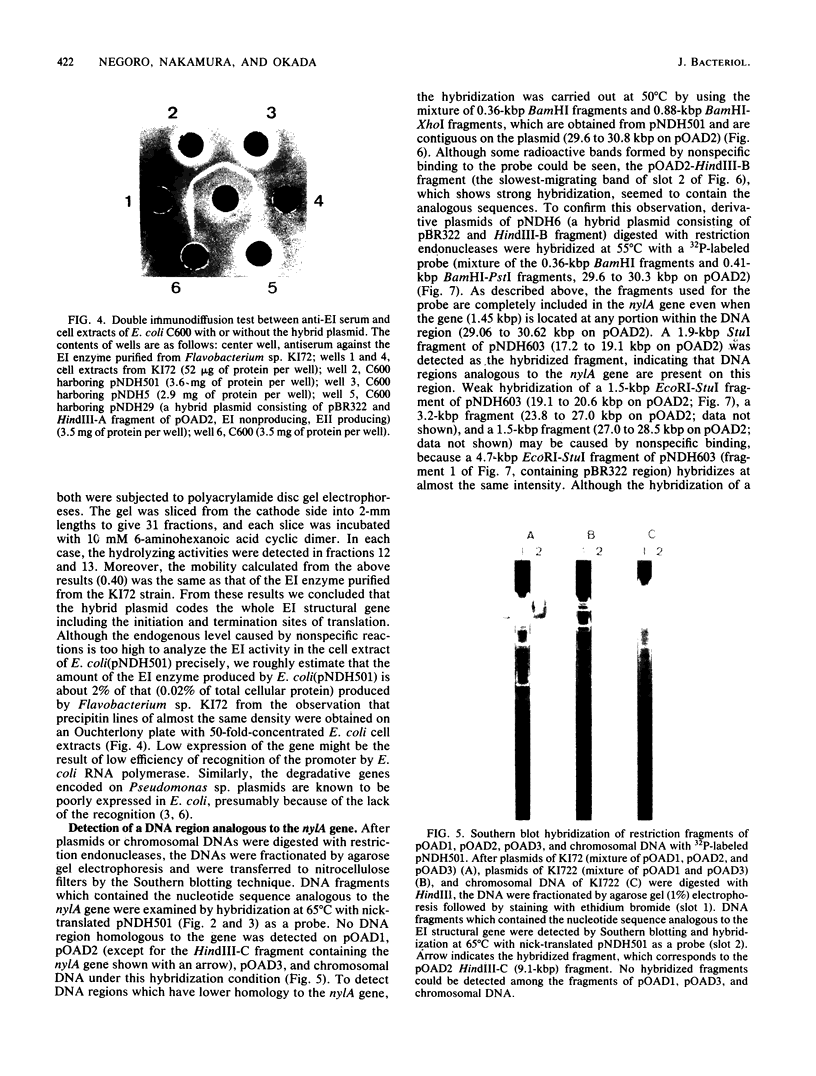
Fine structure of the gene of 6-aminohexanoic acid cyclic dimer hydrolase, one of the enzymes responsible for the degradation of the nylon oligomer (6-aminohexanoic acid cyclic dimer), on the plasmid pOAD2 harbored in Flavobacterium sp. KI72 was determined by constructing miniplasmids from plasmid pNDH5 (a hybrid plasmid consisting of pBR322 and a 9.1-kilobase-pair HindIII fragment of pOAD2 ). The 6-aminohexanoic acid cyclic dimer hydrolase produced by cells of Escherichia coli C600 harboring pNDH5 or its miniplasmid was examined immunologically and electrophoretically and was found to be identical to that of Flavobacterium sp. KI72 . A fragment of pOAD2 (17.2- to 19.1-kilobase-pair region on pOAD2 ) was detected as hybridized fragment by Southern blotting experiments, indicating the presence of the DNA region analogous to the 6-aminohexanoic acid cyclic dimer hydrolase gene on the plasmid.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty A. M., Friello D. A., Bopp L. H. Transposition of plasmid DNA segments specifying hydrocarbon degradation and their expression in various microorganisms. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3109–3112. doi: 10.1073/pnas.75.7.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee D. K., Chakrabarty A. M. Genetic rearrangements in plasmids specifying total degradation of chlorinated benzoic acids. Mol Gen Genet. 1982;188(2):279–285. doi: 10.1007/BF00332688. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Molecular cloning of TOL genes xylB and xylE in Escherichia coli. J Bacteriol. 1981 Mar;145(3):1137–1143. doi: 10.1128/jb.145.3.1137-1143.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita S., Negoro S., Muramatsu M., Bisaria V. S., Sawada S., Okada H. 6-Aminohexanoic acid cyclic dimer hydrolase. A new cyclic amide hydrolase produced by Achromobacter guttatus KI74. Eur J Biochem. 1977 Nov 1;80(2):489–495. doi: 10.1111/j.1432-1033.1977.tb11904.x. [DOI] [PubMed] [Google Scholar]
- Kinoshita S., Terada T., Taniguchi T., Takene Y., Masuda S., Matsunaga N., Okada H. Purification and characterization of 6-aminohexanoic-acid-oligomer hydrolase of Flavobacterium sp. Ki72. Eur J Biochem. 1981 Jun 1;116(3):547–551. doi: 10.1111/j.1432-1033.1981.tb05371.x. [DOI] [PubMed] [Google Scholar]
- Kördel W., Schneider F. Chemical investigations on pig kidney aminoacylase. Biochim Biophys Acta. 1976 Sep 14;445(2):446–457. doi: 10.1016/0005-2744(76)90098-x. [DOI] [PubMed] [Google Scholar]
- Legerski R. J., Hodnett J. L., Gray H. B., Jr Extracellular nucleases of pseudomonas BAL 31. III. Use of the double-strand deoxyriboexonuclease activity as the basis of a convenient method for the mapping of fragments of DNA produced by cleavage with restriction enzymes. Nucleic Acids Res. 1978 May;5(5):1445–1464. doi: 10.1093/nar/5.5.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman I. R. T4 DNA polymerase. Methods Enzymol. 1974;29:46–53. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negoro S., Shinagawa H., Nakata A., Kinoshita S., Hatozaki T., Okada H. Plasmid control of 6-aminohexanoic acid cyclic dimer degradation enzymes of Flavobacterium sp. KI72. J Bacteriol. 1980 Jul;143(1):238–245. doi: 10.1128/jb.143.1.238-245.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negoro S., Taniguchi T., Kanaoka M., Kimura H., Okada H. Plasmid-determined enzymatic degradation of nylon oligomers. J Bacteriol. 1983 Jul;155(1):22–31. doi: 10.1128/jb.155.1.22-31.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H., Negoro S., Kimura H., Nakamura S. Evolutionary adaptation of plasmid-encoded enzymes for degrading nylon oligomers. Nature. 1983 Nov 10;306(5939):203–206. doi: 10.1038/306203a0. [DOI] [PubMed] [Google Scholar]
- Peacock A. C., Dingman C. W. Resolution of multiple ribonucleic acid species by polyacrylamide gel electrophoresis. Biochemistry. 1967 Jun;6(6):1818–1827. doi: 10.1021/bi00858a033. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Roberts T. M., Kacich R., Ptashne M. A general method for maximizing the expression of a cloned gene. Proc Natl Acad Sci U S A. 1979 Feb;76(2):760–764. doi: 10.1073/pnas.76.2.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAITO H., MIURA K. I. PREPARATION OF TRANSFORMING DEOXYRIBONUCLEIC ACID BY PHENOL TREATMENT. Biochim Biophys Acta. 1963 Aug 20;72:619–629. [PubMed] [Google Scholar]