Abstract
There is a vivid debate on the relative importance of local and regional factors in shaping microbial communities, and on whether microbial organisms show a biogeographic signature in their distribution. Taking a metacommunity approach, spatial factors can become important either through dispersal limitation (compare large spatial scales) or mass effects (in case of strongly connected systems). We here analyze two datasets on bacterial communities [characterized by community fingerprinting through denaturing gradient gel electrophoresis (DGGE)] in meso- to eutrophic shallow lakes to investigate the importance of spatial factors at three contrasting scales. Variation partitioning on datasets of both the bacterial communities of 11 shallow lakes that are part of a strongly interconnected and densely packed pond system <1 km apart, three groups of shallow lakes ≈100 km apart, as well as these three groups of shallow lakes combined that span a large part of a North-South gradient in Europe (>2,500 km) shows a strong impact of local environmental factors on bacterial community composition, with a marginal impact of spatial distance. Our results indicate that dispersal is not strongly limiting even at large spatial scales, and that mass effects do not have a strong impact on bacterial communities even in physically connected systems. We suggest that the fast population growth rates of bacteria facilitate efficient species sorting along environmental gradients in bacterial communities over a very broad range of dispersal rates.
Keywords: dispersal limitation, metacommunity biology, microbial biogeography, microbial community, mass effects
Microbial communities may constitute the majority of the earth's biodiversity and catalyze processes that are critical to sustaining life on earth. Understanding the mechanisms that govern their distribution is thus of great interest. There is currently a vivid debate on whether microbial communities share patterns of distribution and diversity similar to those of macroscopic organisms (1–7). The traditional hypothesis among microbiologists, “everything is everywhere, but the environment selects” (8), presumes ubiquity based on the high dispersal rates for microorganisms. This hypothesis has been reinforced by a number of studies in pro- as well as eukaryotic microorganisms that showed the same species or lineage to be present in very different parts of the world (e.g., refs. 9 and 10). However, the results of these studies have been questioned by researchers who claim that insufficient ability to discriminate cryptic taxa may have led to the wrong conclusion that many taxa are cosmopolitan (e.g., ref. 11). In addition, a number of recent studies suggest that some microbial taxa can exhibit geographical isolation and marked biogeographical patterns (3, 12, 13).
Bacterial communities can be locally controlled by a multitude of factors, including habitat size and heterogeneity, ecosystem productivity, biological interactions (competition and predation), and human impact (14–18). If bacteria are ubiquitous because of high dispersal rates, then we expect no differences in community composition in different sites after eliminating the response to environmental variables. This scenario conforms to the model of strong species sorting in a metacommunity framework (19, 20). If, however, bacteria show some dispersal limitation, we expect in addition to the environmental signal a relationship between community composition and location, which may either be due to chance effects or to a biogeographical signal reflecting changes in the regional species pool with distance. Spatial factors may also interfere with the signal of local environmental conditions at the other extreme of the gradient in dispersal rates, however. In very strongly connected habitats, dispersal rates may be so high that they lead to homogenization through mass effects (19). In the metacommunity concept, species sorting results in a matching between the environmental gradients and taxon composition, and is impeded by either too low (dispersal limitation) or too high dispersal rates (mass effects; source-sink dynamics). It follows that it is important to quantify the relative contribution of regional factors (dispersal) and local environmental conditions (species sorting; we here use this term, commonly applied in metacommunity theory, also for bacterial communities, but acknowledge that species delimitation needs other criteria in asexually reproducing prokaryotes than in sexually reproducing taxa) to bacterial community composition over a wide range of spatial scales. To date, only a few studies have investigated the relative influence of geographic distance and habitat factors on bacterial community composition (BCC) (see refs. 6 and 22 for an overview), and none covered an extensive range of spatial scales (e.g., refs. 2, 16, 18, and 21).
Dolan (22), in his review on microbial biogeography, stated that the contrasting patterns and trends observed in different studies may reflect differences in temporal and spatial scales. He concluded that patterns of biogeography are more likely to emerge in studies that focus on similar habitats across different spatial scales. In the present study, we therefore set out to quantify the relative importance of local environmental factors and spatial distance in two datasets on BCCs of the same habitat type, shallow meso- to eutrophic lakes, collected at widely different spatial scales. A first dataset consists of bacterial communities of 98 lakes located in three geographic regions separated by several hundreds to >2,500 km: 32 lakes in Denmark (DK), 34 in The Netherlands and Belgium (BNL), and 32 in southern Spain (SP) [supporting information (SI) Fig. 2]. This dataset is analyzed at both the regional and the near-continental level. At the regional scale, lakes were on average in DK 23 km separated from each other (range 1–78 km distance among individual lakes), in BNL 97 km (range 0.1–250 km), and in SP 185 km (range 0.2–430 km). The most distant lakes in the combined area DK and BNL are separated 1,060 km; the most distant lakes in the total dataset are separated by 3,100 km. The second dataset consists of bacterial communities of 11 lakes in a strongly interconnected pond system that encompasses 34 small shallow lakes on a total area of <300 ha (SI Fig. 3) (23). We used a 16S rRNA gene-based fingerprinting technique, denaturing gradient gel electrophoresis (DGGE), to determine BCC. The fingerprints consisted of banding patterns, where each band was translated to one operational taxonomic unit (OTU) that was considered as a surrogate of the predominant bacterial “species” present. Our specific goals were to (i test to which degree geographic distance has an influence on BCC in these two widely contrasting datasets focusing on the same and common habitat category, and (ii) identify the (environmental; spatial) factors that best explain variation in BCC. We take a metacommunity approach (19) as a logical framework to study the impact of local and regional factors.
Results
European Dataset.
A total of 107 different operational taxonomic units (OTUs) were detected from the 98 study lakes. Eighty-eight OTUs were recorded in DK, 86 in BNL, and 98 in Spain. The total number of OTUs found in one lake ranged from 11 to 37. Although 85% of the OTUs (94 of 107) were detected in all three geographic regions, there was a clear overall differentiation in BCC between these regions, confirmed by an analysis of similarities (ANOSIM) test (r = 0.18, P < 0.001). Pairwise tests revealed significant differences between DK and SP (r = 0.22, P < 0.001), between BNL and SP (r = 0.24, P < 0.001), and between DK and BNL (r = 0.12, P < 0.001). The dissimilarity in BCC tended to be lower between DK and BNL (average dissimilarity: 50%) than between DK and SP (60%), or between BNL and SP (59%).
The results of the similarity percentage (SIMPER) analyses of the transformed abundance identifying the OTUs that contribute most strongly to the dissimilarity between geographic regions are given in SI Table 1; parts of these bands were excised and sequenced. The Spanish samples show a higher average abundance of a member of the Actinomycetes, subgroup Agrococcus jenesis (DGGE 67.4), a member of the Bacteroidetes subgroup CL500-6 (DGGE 51.1), and an Aeromonas like organism (DGGE 65.8). Conversely, in the DK and BNL lakes, we found a higher average abundance of members of the Actinomycetes subgroup ACK-M1 (DGGE 79.4, 59.8, 52.5, 63.2), Bacteroidetes Cytophaga subgroup (DGGE 39.2), Phormidium limnetica (DGGE 37.3), and Alphaproteobacteria LD12 (DGGE 32.9). Bacterioplankton communities in BNL differed from DK mainly because of higher abundance of a member of the alpha-proteobacteria (DGGE: 32.9) and a member of the CFB-group (DGGE: 42.2), and a lower abundance of a member of the genus Synechococcus (DGGE: 41.5). Some OTUs showed an abundance gradient from North to South (increasing: DGGE 79.4, 52.5, 39.2, 20.3, 58.0; decreasing: DGGE 50.1, 20.3, 47.8, 65.8, 67.4).
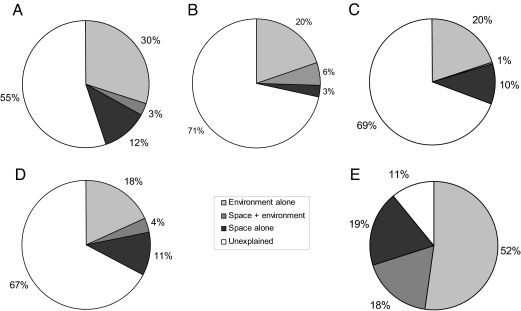
Considering the three regions together in one analysis, variation partitioning between significant environmental variables and spatial location (here limited to a grouping variable indicating what region the lake belongs to) yielded 3% (1% unbiased) of total variance explained by region, 20% (8%) by environmental variables, and 6% (6%) by a common environment-region effect (Fig. 1A and SI Table 2). A large amount of variation (71%, 85% unbiased) remains unexplained in this dataset. The environmental variables that significantly contributed to explain the overall BCC patterns after removal of the region effect were: depth of the lake, pH, total nitrogen concentration (TN), bacterial densities, % Bosmina, densities of heterotrophic nanoflagellates (HNFDENS), biomass of ciliates, % grassland, and % forest (SI Fig. 4A).
Fig. 1.
Variation partitioning of the BCC. Shown are the three regions together (A), Denmark (B), Belgium and the Netherlands (C), Spain (D), and “De Maten” (E).
Restricting our analysis to DK only, environmental and spatial variables (third order polynomials of projected longitudes and latitudes) explained 31% (10% unbiased) of the total variance. Twenty percent (9% unbiased) of the total variance was explained by pure environmental variables. After removal of environment-related variation, space did no longer significantly explain any variation in the data collected in DK (Fig. 1B and SI Table 2). The environmental variables that significantly explained BCC patterns in the DK lakes were as follows: lake area, TN, biomass of Bosmina, total zooplankton (ZOOPTOT) and Ceriodaphnia, % Cyanobacteria, and % ultraphytoplankton (SI Fig. 4B).
For the BNL data, variance partitioning between significant environmental variables and spatial variables yielded 12% (6% unbiased) of total variance explained by pure spatial variables, 30% (15%) by environmental variables and 3% (1%) by common effects (Fig. 1C and SI Table 2). The amount of variation not explained by the environmental and spatial variables was 55% (79%). The environmental variables that significantly explained BCC patterns were as follows: lake area, TN, biomass of Bosmina, ZOOPTOT, and Ceriodaphnia, % Cyanobacteria, and % ultraphytoplankton (SI Fig. 4C). This list of environmental variables that significantly explain variance in BCC in the BNL lakes is identical to that of the DK lakes.
For SP, variance partitioning between significant environmental variables and spatial variables yielded 11% (3% unbiased) of total variance explained by pure spatial variables, 18% (8%) by environmental variables, and 4% (2%) by common effects (Fig. 1D and SI Table 2). A large amount of variation (67%, 87% unbiased) was not explained by the environmental and spatial variables. The environmental variables that significantly explained the BCC patterns were as follows: depth, temperature, HNFDENS, and total coverage of submerged macrophytes (SMTOT) (SI Fig. 4D).
Because of the overlap in environmental variables explaining BCC in BNL and DK, we explored this similarity in detail. First, we calculated Pearson correlations between the relative abundance of taxa and all environmental factors. We observed that for 10 of the 55 taxa (18%), significant correlations were observed with the same environmental variables or combination of environmental variables (SI Table 3). This number increased to 17 (31%) when we relaxed significance criteria of the correlations to 0.1. This pattern was confirmed by our observation that similarity matrices among taxa for their correlations with environmental variables were significantly related between the two regions (Relate function in Primer; rho = 0.082; P = 0.005).
De Maten Dataset.
A total of 42 different OTUs were detected in the 11 lakes of the strongly connected De Maten system. The number of OTUs per lake varied from 16 to 23. Seven OTUs occurred in all lakes, and represented on average 47% (33–68%) of the relative band intensity in the lakes. The average dissimilarity between the lakes was relatively low (40.7%) and varied between 56% (between lakes 12 and 17) and 11% (between lakes 9 and 10). The turbid lakes 9, 10, 11, and 12 had a similarity in BCC exceeding 80%; their high similarity was mainly due to the presence of the same dominant OTU that made up 21–30% of the relative band intensity and was much less intense in the other lakes (< 6% relative band intensity).
Variance partitioning on this dataset revealed that environmental and spatial variables explained 89% (46% unbiased) of the total variance. Fifty-two percent (36%) of the total variance was explained by pure environmental variables. After removal of environment-related variation, spatial configuration did no longer significantly explain any variation (Fig. 1E and SI Table 2). The environmental variables that significantly explained the BCC pattern in the De Maten lakes were transparency (Sneller depth), pH, conductivity, biomass of dinoflagellates, and biomass of calanoid copepods (SI Fig. 4E).
Discussion
At the largest spatial scale, we observed a differentiation in BCC among geographic regions. This differentiation is strongest between the Spanish lakes and the lakes of the more northern regions (DK and BNL). There are several factors that may account for this among-region effect. One possible explanation is that regional differences in BCC reflect the biogeography of bacteria, implying that there would be a certain amount of historical contingency impacting bacterial distribution patterns, similar to the patterns observed for multicellular organisms. Rather than every species potentially being everywhere, the array of taxa that may colonize a given patch would then be confined to a more regional taxon pool. Alternatively, the patterns observed in our study may reflect regional differences in environmental conditions, either not measured in our study or confounded in spatial differences. Several lines of evidence suggest that the latter explanation is more likely than a biogeography mediated by dispersal limitation. First, as >85% of the detected bacterial OTUs were found in all three studied regions, we can conclude that the majority of freshwater bacterial taxa are not confined to a subset of regions at this geographical scale (<3,000 km). It is therefore unlikely that a strong biogeography effect plays an important role in determining the BCC at a given location. Second, when environmental variables were taken into account, geographic distance alone explained only a very low percentage of the total variation (3%). Our data thus suggest that lakes with similar environmental characteristics have similar bacterial communities regardless of geographic distance. This observation is strongly supported when we focus on BNL and DK only, two regions for which the lakes in general are ecologically more similar than when the Spanish lakes are included (24). For the datasets of DK and BNL, the same set of environmental variables is selected as significantly impacting BCC in local lakes. Our observation that the pattern of correlations of the relative abundance of almost 20% of the taxa is significantly related among these two regions suggests that a significant part of the BCC of lakes in the combined area (covering a distance of >1,000 km) is structured in a similar way by the environment. Given that our analysis captures only part of the relevant environmental factors (e.g., not including sources of DOM, the major food source of the bacteria), this observation is striking. Overall, our data thus suggest that the two regions basically share the same regional taxon pool. The BCC of the lakes in SP did show appreciable differentiation from that of DK and BNL, but the same holds for the ecology of the lakes (24).
Our observations on the wide distribution of bacterial taxa are in agreement with the finding of Yannarell and Triplett (16), who observed differences in bacterial communities between northern and southern Wisconsin lakes, but also found that most of the bacterial taxa were distributed state-wide. Fierer and Jackson (25) came to the same conclusion in their study on the biogeography of soil bacterial communities, covering 98 soil samples from across North and South America. These studies and our study suggest that the distribution of bacterial taxa is not strongly limited by dispersal even at relatively large spatial scales [up to several thousands of km, i.e., spatial scales at which the influence of both historical contingencies and contemporary ecological factors on microbial biogeography are most likely to be detected (6)]. Our results further agree with Horner-Devine et al. (2), who found that the taxa-area relationship for bacteria in salt marsh sediments was driven primarily by environmental heterogeneity, which increased with increased area considered, rather than by geographic distance itself.
Dolan (22) suggested that a signature of biogeography would more likely emerge in studies that focus on similar habitats across space. A key feature of our study is that similar lake types were selected across all spatial scales. All studied lakes were shallow and meso- to eutrophic, and they were selected to fit into 16 categories comprising all combinations of large/small, connected/isolated, relatively low versus relatively high potential productivity, and absence/presence of macrophytes in the three regions studied. This sampling design ensured that very similar lakes were selected in the different regions, although this strategy worked better for BNL and DK than for SP (24). Even though this sampling design made our study ideally suited to detect a signature of biogeography (22), no strong effect of geographic distance was detected. Although there are some recent reports demonstrating that dispersal limitation may occur at large spatial scales for some microbial organisms (12), the environments involved were extreme and isolated, and may thus perhaps not be representative of typical and common surface water categories (rivers and rivulets, ditches, ponds, and lakes). In rare and more isolated habitat types, one is more likely to detect dispersal limitation, as dispersal rates are also a function of the amount and size of source populations. Our results are indeed not incompatible with the idea that a subset of bacterial taxa may be more strongly dispersal limited. Rather, they suggest that the majority of taxa present in natural bacterial communities seem to be very widely distributed.
At the other extreme of the spatial spectrum, we show a much higher association of BCCs with environmental factors in the studied interconnected pond system than with spatial factors. These results are in contrast with Reche et al. (21), who found that the location of water bodies in the Sierra Nevada influenced bacterial communities and that BCCs of nearby lakes showed more similar OTU compositions. They argued that this pattern is due to recolonization of a lake by microorganisms from adjacent lakes being more frequent than from more remote lakes. Similarly, Lindström et al. (18) showed that BCC of lakes with a residence time <100 days showed a signature of mass effects, reflected by similarity of BCC of the lake community with that of the inlet. The discrepancy between these studies and ours may lie in the fact that we studied meso- to eutrophic systems, in which species replacements are expected to occur faster (i.e., more efficient species sorting) than in oligotrophic systems.
In the framework of metacommunity theory (19), dispersal rates play a key role in determining patterns of community similarities among habitat patches. Very low dispersal rates can lead to dispersal limitation, resulting in purely spatial biogeography patterns. Very high dispersal rates can lead to mass effects, with taxa also occurring in less suitable habitats because of continuous supply. Mass effects thus also lead to purely spatial effects and a reduction of the match between the occurrence of specific OTUs and environmental conditions. Intermediate dispersal rates provide the best conditions for environmental factors to determine community composition, as there is a sufficient supply of taxa from the regional species pool to fuel species sorting, whereas there is no blurring of the resulting differences in community composition by mass effects. In this gradient of environmental connectedness, the impact of environment is thus highest at intermediate dispersal levels. In the case of mass effects, one may anticipate that the impact of spatial factors is more pronounced at relatively small spatial scales, whereas the impact of spatial factors is expected to increase with geographical scale in the case of dispersal limitation. Our data suggest that BCCs comply to the scenario of species sorting, suggesting intermediate dispersal rates when scaled to the efficiency of species sorting along environmental gradients. Intriguingly, our data suggest that this strong species sorting occurs at the near-continental (European) scale as well as at the within-regional and local scale. This finding suggests that dispersal rates at a scale of >1,000 km are still for most taxa high enough to allow species sorting to lead to a good association between BCC and environmental variation, while at the same time dispersal rates at a local scale of interconnected systems are not so high as to result in strong mass effects. At the intermediate scale, a very high impact of species sorting is observed. In the region with the smallest inter-lake distances (DK), the impact of spatial factors was indeed not significant when environmental differences are taken into consideration. Importantly, in the above, the dispersal gradient has to be viewed relative to the strength and rates of species sorting. Indeed, if species sorting is very efficient and rapid, dispersal rates must be very high to result in mass effects (e.g., ref. 26). It follows that our data strongly suggest that species sorting in bacterial communities is very efficient, at least in the meso- to eutrophic systems studied, so that bacterial communities track environmental conditions even in the presence of very high immigration rates of bacterial taxa from other sources. Similarly, their very high population growth rates make BCCs largely independent of the amount of propagules that arrive in the focal habitat, extending the range of actual dispersal rates over which species sorting can strongly impact BCC to the lower end of the spectrum. Our results thus suggest that species sorting in BCCs is very efficient at both very low and very high dispersal rates. This efficiency of space sorting is most likely due to the extremely high population growth rates of bacteria. Several studies that focused on the dynamics of BCCs in local habitats have indeed reported that bacterial communities can rapidly track changes in the environment (14, 27–31).
Conclusion
Our results provide strong evidence that species sorting in response to local environmental factors is a key determinant of the taxon composition of aquatic bacterial communities over a very broad range of spatial scales (<100 m to >1,000 km). The main factors controlling bacterial community composition were resources (TN) and grazing-related factors (e.g., zooplankton biomass). We argue that species sorting is so important in bacterial communities because the high population growth rates of bacteria largely uncouple local population dynamics and relative abundances of taxa from dispersal rates. In combination with sufficiently high dispersal rates to allow colonization of distant habitats, these high population growth rates lead to a weak biogeographical signal. Bacterial taxa need not be everywhere at all times to yield the observed pattern: it is sufficient that low but sustained or regular dispersal is coupled with very efficient tracking of environmental conditions through local population dynamics. The high population growth rates of microbial organisms in this way result in a much broader range of spatial scales over which species sorting plays a predominant role in shaping community structure than in many macroorganisms. The power of species sorting in microbial communities thus does not only reflect high dispersal rates, but rather the interplay of sufficiently high dispersal rates (caused by small size, production of resistant stages, and vast population sizes) and efficient tracking of environmental changes due to fast population growth rates. This framework explains metacommunity structure of microorganisms using the same mechanisms as in macroorganisms, and has in our opinion the potential to reconcile the two opposing viewpoints on the occurrence of biogeographical signals in microbial communities. It predicts that the biogeographical signal for many microorganisms is much weaker than for most macroorganisms because of the fact that long distance dispersal is more likely combined with a strong capacity to establish viable populations when environmental conditions allow. It also predicts that biogeographical signal is likely to be stronger in microorganisms inhabiting rare habitats. Both predictions are in line with current observations (e.g., ref. 12).
Materials and Methods
Selection and Sampling of Lakes.
European scale.
Ninety-eight shallow lakes were sampled, located in three European regions at different latitudes: Denmark (32 lakes), Belgium/The Netherlands (34 lakes), and southern Spain (32 lakes) (SI Fig. 2). Lakes were not selected randomly, but according to four potentially important key factors: submerged vegetation cover (more than or less than 20% of the lake area surface covered), total phosphorus (more than or less than 100 μg·l−1), lake surface area (more than or less than 5 ha), and the degree of connectedness (“isolated”: distance to nearest lake longer than 200 m; first order lake if part of river system; “connected”: distance to nearest lake <200 m; second or higher order lake and distance to upstream lake smaller than 1 km if part of river system; see refs. 24 and 32). For a detailed description of the sampled lakes, see Declerck et al. (24).
Each lake was sampled monthly during a period of 6 months in summer, either in 2000 or 2001. To integrate spatial variability within each lake, samples were collected at 8 (lakes <5 ha) or 16 (lakes >5 ha) randomly selected locations and pooled. To obtain DNA samples of bacteria, 5 liters of the pooled sample was fractionated by using a mesh of 20 μm, separating bacteria that were free-living or attached to small seston particles from organisms attached to large particles. The small fraction was then filtered over a 0.2-μm MF-Millipore MCE filter and kept frozen (−80°C) until further analysis.
Local scale: The pond system “De Maten.”
The De Maten wetland consists of a series of 34 interconnected shallow lakes that are located within a small geographic area (≈300 ha). They are fed mainly by two rivulets and are connected by a system of rivulets and overflows (SI Fig. 3). As a result, connectivity in this system is extremely high. Although nutrient levels and water chemistry are similar among the lakes in this system, they do differ strongly in their ecology, as some lakes are turbid whereas others are in a clear-water state with a dense vegetation of submerged macrophytes. For more detailed information on the environmental conditions in these lakes, see Cottenie et al. (23, 33). Information on the morphometry and water flow between the lakes is given in Michels et al. (34, 35). Eleven lakes of this system were sampled in August 2001. The lakes could not be selected according to the gradients used in the European-scale study, as there is relatively little variation in connectivity, size, and TP concentration in this set of lakes. The 11 lakes were therefore chosen to be centrally located in the system (SI Fig. 3) and to consist of both lakes in the turbid and in the clear-water state (i.e., absence/presence of dense macrophyte vegetation). More information on the measured environmental variables is given in SI Text.
Analysis of Samples.
DNA extraction and PCR.
DNA was extracted directly by using the bead-beating method concomitant with phenol extraction and ethanol precipitation (36). After extraction, the DNA was purified on a Wizard column (Promega, Madison, WI) according to the manufacturer's recommendations. After DNA extraction and purification, the DNA of the six monthly samples of the lakes in the European study was pooled so that we obtained one sample per lake; for the De Maten lakes, only one sample was taken per lake (August 2001).
For DGGE analysis, a small rDNA fragment was amplified with primers 357F-GC-clamp (5′-CGCCCGCCGCGCCCCGCGCCCGGCCCGCCGCCCCCGCCCCCCTACGGGAGGCAGCAG-3′) and 518R (5′-ATTACCGCGGCTGCTGG-3′). By using these specific primers, our analysis is restricted to the domain Bacteria. The PCR mixture containing 5 μl of template DNA, each primer at a concentration of 0.5 μM, each deoxynucleosidetriphosphate at a concentration of 200 μM, 1.5 mM MgCl2, 20 ng of BSA, 5 μl of 10× PCR buffer [100 mM Tris·HCl (pH 9), 500 mM KCL], and 2.5 units of TaqDNA polymerase (Ampli-Taq PerkinElmer) was adjusted to a final volume of 50 μl with sterile water (Sigma). After an incubation for 5 min at 94°C, a touchdown PCR was performed by using 20 cycles consisting of denaturation at 94°C for 1 min, annealing at 65°C (initial cycle, the temperature was decreased by 0.5°C every cycle) for 1 min and primer extension at 72°C for 1 min. Five additional cycles were carried out at an annealing temperature of 55°C. The tubes were then incubated for 10 min at 72°C. The concentration of PCR products was determined by analyzing 5 μl of product on 1% (wt/vol) agarose gels, staining with ethidium bromide, and comparison with a molecular weight marker (Smartladder; Eurogentec).
Analysis of BCC by denaturing gradient gel electrophoresis (DGGE).
PCR products were analyzed on a 35–70% denaturant DGGE gel as described in ref. 27. DGGE gels were stained with ethidium bromide and photographed on a UV transillumination table (302 nm) with a CCD camera. The 98 samples of the European lakes were randomly analyzed on nine parallel DGGE-gels; all samples of “De Maten” lakes were analyzed on one gel. As standards, we used a mixture of DNA from nine clones, obtained from a clone library of the 16S rRNA genes from Lake Visvijver (27). On every gel, three standard lanes were analyzed in parallel to the samples. The position of the bands in the standard lanes was used to compare the patterns formed in different gels. Digitized DGGE images were analyzed by using the software package Bionumerics 1.5. (Applied Maths BVBA, Kortrijk, Belgium). The software performs a density profile through each lane, detects the bands, and calculates the relative contribution of each band to the total band signal in the lane after applying a rolling disk as background subtraction. Bands occupying the same position in the different lanes of the gel were first identified by the program and then visually checked. After alignment within each gel, the gels were aligned against each other using the standards. After two-by-two alignment, verification alignment of other combinations of gels proved straightforward and repeatable. Samples loaded on more than one gel were used as an extra quality check. Finally, a matrix was compiled based upon the relative contribution of individual bands to the total band signal in each lane.
Nucleotide sequences of DGGE bands of interest were obtained by direct sequencing of DNA from excised DGGE bands as described in ref. 27. Sequencing was performed with the ABI-Prism sequencing kit (PE-Biosystems) using the primer R519 (5′-GTATTACCGCGGCTGCTG-3′) and an automated sequencer (ABI-Prism 377). In total, 104 bands were cut out, 60 of which yielded nice sequences. A GenBank Blast search was performed for each of those sequences to identify the closest relatives. The partial sequences obtained in this study have been deposited in the GenBank database under accession numbers AM748764–AM748785. The occurrence of chloroplasts in our samples was low (3 of 60 verified sequences).
Data Analysis.
For both datasets separately, we determined the relative importance of environmental characteristics and spatial processes in explaining differentiation in BCC by decomposing the total variation in the bacterial community matrix into unique environmental and spatial components with corresponding P values using (partial) redundancy analysis (23, 37, 38). This multivariate extension of linear regression with corresponding R2 measures the amount of variation (computed as the percentage of the total variation in the community matrix) that can be attributed exclusively to one or the other set of explanatory environmental (E) or spatial (S) variables. The different components are as follows: total explained variation [E+S], environmental variation [E], spatial variation [S], environmental variation without a spatial component [E/S], and spatial variation without an environmental component [S/E]. The significance of these components was evaluated with a Monte Carlo permutation test (999 new values under the null hypothesis). For the partial RDA analyses [E/S] and [S/E], residuals under the “reduced” model were permuted; for the other RDA analyses ([E+S], [E], and [S]), residuals under the “full” model were permuted (38). We computed two other fractions: (i) the unexplained variation (1 − [E+S]) and (ii) the variation explained by correlations between environmental and spatial variables ([E with S] = [E] − [E/S] = [S] − [S/E]). Peres-Neto et al. (39) showed that these variation components are biased estimates and provided formulas for unbiased estimates. We reported both the original (38) and the new unbiased estimates of the explained variation components (39) to ease comparisons with studies that have not incorporated this recently discovered method. BCC data were fourth-root transformed to normalize these skewed density data. At the European scale, spatial variables measured as latitude–longitude coordinates were converted into projected coordinates. For the “De Maten” system, we used the coordinates based on effective dispersal distances between lakes that incorporate the dispersal pathways between the different lakes as quantified in a GIS model parametrized using field measurements (34, 35). All third-order polynomials of the projected coordinate variables were constructed to capture more complicated spatial patterns (38). For the environmental variables of the European lakes, we used averages calculated from the six monthly measurements for abiotic environmental variables and chlorophyll a, and worked with count data from pooled samples (equal volume) for biotic variables such as zooplankton. Macrophyte cover was quantified once at the height of the growing season using a standardized survey. Because increasing the number of explanatory variables results in an increase of explained variation, we limited both the environmental and spatial third degree polynomial variables to the most parsimonious subset of significant variables each with a forward selection procedure. This procedure also eliminated overfitting and problems with colinear variables. R (40) and the Vegan library (41) were used for the RDA analyses.
For the European dataset, we also determined whether the three regions differ in overall community composition with an analysis of similarities (ANOSIM; refs. 42 and 43) and identified which species are associated with the different regions with a similarity percentage (SIMPER; ref. 43) analysis. Both analyses were performed in PRIMER 5 for Windows (44).
Supplementary Material
ACKNOWLEDGMENTS.
We thank the many people who helped in sampling and quantifying ecological characteristics of the study lakes. We cordially thank all lake owners and managers for allowing access to the lakes. S.D. is a postdoctoral fellow of FWO-Vlaanderen. This study was funded by European Union-projects BIOMAN (Grant EVK2-CT-1999-00046) and EURO-LIMPACS (Grant GOCE-CT-2003-505540) and by European Science Foundation EURODIVERSITY project BIOPOOL (cofunded by FWO and Belspo).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AM748764–AM748785).
This article contains supporting information online at www.pnas.org/cgi/content/full/0707200104/DC1.
References
- 1.Fenchel T. Science. 2003;301:925–926. doi: 10.1126/science.1089242. [DOI] [PubMed] [Google Scholar]
- 2.Horner-Devine MC, Carney KM, Bohannan BJM. Proc R Soc London Ser B. 2004;271:113–122. doi: 10.1098/rspb.2003.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell T, Ager D, Song J-I, Newman JA, Thompson IP, Lilley AK, vander Gast CJ. Science. 2005;308:1884. doi: 10.1126/science.1111318. [DOI] [PubMed] [Google Scholar]
- 4.Fenchel T, Finlay BJ. Science. 2005;309:1995. doi: 10.1126/science.309.5743.1997. [DOI] [PubMed] [Google Scholar]
- 5.Mitchell EAD. Science. 2005;309:1995. [Google Scholar]
- 6.Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR, et al. Nat Rev. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- 7.Dolan JR. J Biogeogr. 2006;33:199–200. [Google Scholar]
- 8.Baas-Becking LGM. Geologie of Inleidning tot de Milieukunde. The Hague, The Netherlands: W P Van Stokum; 1934. [Google Scholar]
- 9.Finlay BJ, Clarke KJ. Nature. 1999;400:828. [Google Scholar]
- 10.Finlay BJ. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- 11.Foissner W. Acta Protozool. 2006;45:111–136. [Google Scholar]
- 12.Whitaker RJD, Grogan W, Taylor JW. Science. 2003;301:976–978. doi: 10.1126/science.1086909. [DOI] [PubMed] [Google Scholar]
- 13.Papke RT, Ward DM. FEMS Microbiol Ecol. 2004;48:293–303. doi: 10.1016/j.femsec.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 14.Jardillier L, Boucher D, Personnic S, Jacquet S, Thénot A, Sargos S, Amblard C, Debroas D. FEMS Microbiol Ecol. 2005;53:429–443. doi: 10.1016/j.femsec.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 15.Van der Gucht K, Vandekerckhove T, Vloemans N, Cousin S, Muylaert K, Sabbe K, Gillis M, Declerck S, De Meester L, Vyverman W. FEMS Microbiol Ecol. 2005;53:205–220. doi: 10.1016/j.femsec.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Yannarell AC, Triplett EW. Appl Environ Microbiol. 2005;71:227–239. doi: 10.1128/AEM.71.1.227-239.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindström ES, Kamst-van Agterveld MP, Zwart G. Appl Environ Microbiol. 2005;71:8201–8206. doi: 10.1128/AEM.71.12.8201-8206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindström ES, Forslund M, Bergström AK. Limnol Oceanogr. 2006;51:339–342. [Google Scholar]
- 19.Leibold MA, Norberg J. Limnol Oceanogr. 2004;49:1278–1289. [Google Scholar]
- 20.Cottenie K. Ecol Lett. 2005;8:1175–1182. doi: 10.1111/j.1461-0248.2005.00820.x. [DOI] [PubMed] [Google Scholar]
- 21.Reche I, Pulido-Villena E, Morales-Baquero R, Casamayor EO. Ecology. 2005;86:1715–1722. [Google Scholar]
- 22.Dolan JR. Aquat Microb Ecol. 2005;41:39–48. [Google Scholar]
- 23.Cottenie K, Michels E, Nuytten N, De Meester L. Ecology. 2003;84:991–1000. [Google Scholar]
- 24.Declerck S, Vandekerkhove J, Johansson L, Muylaert K, Conde-Porcuna JM, Van der Gucht K, Pérez-Martynez C, Lauridsen T, Schwenk K, Zwart G, et al. Ecology. 2005;86:1905–1915. [Google Scholar]
- 25.Fierer N, Jackson RB. Proc Natl Acad Sci USA. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cottenie K, De Meester L. Ecology. 2004;85:114–119. [Google Scholar]
- 27.Van der Gucht K, Sabbe K, De Meester L, Vloemans N, Zwart G, Gillis M, Vyverman W. Environ Microbiol. 2001;3:680–690. doi: 10.1046/j.1462-2920.2001.00242.x. [DOI] [PubMed] [Google Scholar]
- 28.Muylaert K, Van der Gucht K, Vloemans N, De Meester L, Gillis M, Vyverman W. Appl Environ Microbiol. 2002;68:4740–4750. doi: 10.1128/AEM.68.10.4740-4750.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jardillier L, Basset M, Domaizon I, Belan A, Amblard C, Richardot M, Debroas D. Aquat Microb Ecol. 2004;35:259–273. [Google Scholar]
- 30.Kirchman DL, Dittel AI, Findlay SEG, Fischer D. Aquat Microb Ecol. 2004;35:243–257. [Google Scholar]
- 31.Pinhassi J, Sala MM, Havskum H, Peters F, Guadayol O, Malits A, Marrasé C. Appl Environ Microbiol. 2004;70:6753–6766. doi: 10.1128/AEM.70.11.6753-6766.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Meester L, Declerck S, Janse JH, Dagevos JJ, Portielje R, Lammens E, Jeppesen E, Lauridsen T, Schwenk K, Muylaert K, et al. Ecol Stud. 2006;191:150–167. [Google Scholar]
- 33.Cottenie K, Nuytten N, Michels E, De Meester L. Hydrobiologia. 2001;442:339–350. [Google Scholar]
- 34.Michels E, Cottenie K, Neys L, De Meester L. Hydrobiologia. 2001;442:117–126. [Google Scholar]
- 35.Michels E, Cottenie K, Neys L, De Gelas K, Coppin P, De Meester L. Mol Ecol. 2001;10:1929–1938. doi: 10.1046/j.1365-294x.2001.01340.x. [DOI] [PubMed] [Google Scholar]
- 36.Zwart G, Huismans R, van Agterveld MP, Van de Peer Y, De Rijk P, Eenhoorn H, Muyzer G, van Hannen EJ, Gons HJ, Laanbroek HJ. FEMS Microbiol Ecol. 1998;25:159–169. [Google Scholar]
- 37.Borcard D, Legendre P, Drapeau P. Ecology. 1992;73:1045–1055. [Google Scholar]
- 38.Legendre L, Legendre P. Numerical Ecology. New York: Elsevier; 1998. [Google Scholar]
- 39.Peres-Neto PR, Legendre P, Dray S, Borcard D. Ecology. 2006;87:2614–2625. doi: 10.1890/0012-9658(2006)87[2614:vposdm]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 40.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2007. [Google Scholar]
- 41.Oksanen J, Kindt R, Legendre P, O'Hara RB. Vegan: Community Ecology Package. 2007 Version 1.8–6, http://cran.r-project.org. [Google Scholar]
- 42.Clarke KR. J Ecol. 1993;18:117–143. [Google Scholar]
- 43.Clarke KR, Warwick RM. Change in Marine Communities: An Approach to Statistical Analysis and Interpretation. Plymouth, UK: National Environment Research Council; 1994. [Google Scholar]
- 44.Clarke KR, Gorley RN. PRIMER 5 for Windows. Plymouth, UK: PRIMER-E Ltd.; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.