Abstract
The number of sensory receptor genes varies extensively among different mammalian species. This variation is believed to be caused partly by physiological requirements of animals and partly by genomic drift due to random duplication and deletion of genes. If the contribution of genomic drift is substantial, each species should contain a significant amount of copy number variation (CNV). We therefore investigated CNVs in sensory receptor genes among 270 healthy humans by using published CNV data. The results indicated that olfactory receptor (OR), taste receptor type 2, and vomeronasal receptor type 1 genes show a high level of intraspecific CNVs. In particular, >30% of the ≈800 OR gene loci in humans were polymorphic with respect to copy number, and two randomly chosen individuals showed a copy number difference of ≈11 in functional OR genes on average. There was no significant difference in the amount of CNVs between functional and nonfunctional OR genes. Because pseudogenes are expected to evolve in a neutral fashion, this observation suggests that functional OR genes also have evolved in a similar manner with respect to copy number change. In addition, we found that the evolutionary change of copy number of OR genes approximately follows the Gaussian process in probability theory, and the copy number divergence between populations has increased with evolutionary time. We therefore conclude that genomic drift plays an important role for generating intra- and interspecific CNVs of sensory receptor genes. Similar results were obtained when all annotated genes were analyzed.
Keywords: birth–death process, copy number evolution, human evolution, multigene family, olfactory receptor
Eukaryotic genomes contain many multigene families, and the number of gene copies in a multigene family often varies with organism (1, 2). In particular, the copy numbers of sensory receptor gene families are known to vary extensively among different mammalian species (3–6). A certain portion of this variation is likely to be accounted for by physiological requirements for the species to adapt to their specific environments. However, because these gene families contain a large number of pseudogenes (3–6), they also appear to have experienced a random change of gene copy number caused by duplication, deletion, and inactivation of genes. This random change of copy number during evolution is called genomic drift (7).
If this view is right, we would expect that each species contains a substantial amount of copy number variation (CNV), particularly with respect to sensory receptor genes. That is, different individuals in a species are expected to have different numbers of sensory receptor genes. Fortunately, this problem can now be studied, because a number of authors have studied the copy number variable regions (CNVRs) of the human genome, examining many different individuals from various populations (8–14).
The most extensive study so far conducted is that of Redon et al. (13). They searched for CNVRs exhaustively and studied the copy number polymorphisms of genic and nongenic nucleotide sequences among 270 healthy individuals sampled from Africans (30 parent–offspring trios from Yoruba, Nigeria), Asians (45 unrelated Han Chinese from Beijing, China, and 45 unrelated Japanese from Tokyo, Japan), and Europeans (30 parent–offspring trios of European descents from Utah). Here, a CNVR is defined as a genomic region of a few kilobases to a few megabases, in which different individuals contain different numbers of copies of DNA pieces. Because they used DNA hybridization techniques, the detection of CNVRs was not always accurate, but according to their quality assessment, the false-positive rate for detecting CNVRs was low (≤5%), and they identified 1,447 CNVRs including X and Y chromosomes. These CNVRs include genes that are polymorphic with respect to copy number. In this article, these genes will be called copy number polymorphic genes (CNPGs). Using the gene function annotation from the gene ontology (GO) database (15), Redon et al. (13) examined CNVs for various genes, but GO analysis was quite crude, because the same type of gene was included in several different categories. In the case of sensory receptor genes, however, this problem can be avoided, because the nucleotide sequences of the genes are already known (16–18), and all of the genes within CNVRs can be identified.
In their study of CNVs, Redon et al. (13) used a particular European individual as the reference and examined CNVs between this individual and other sampled individuals. This experimental procedure was used, because the currently available human genome sequence (standard genome sequence) is a mixture of genomes from different individuals. Because the exact number of gene copies in the reference individual is not known, the absolute number of gene copies of each sampled individual cannot be determined. All we can do is to know the copy number relative to that of the reference individual. Yet, the distribution of this number among sampled individuals should be the same as that of the absolute number. In reality, the CNVs determined by Redon et al. (13) were those not among different genomes but among different diploid individuals. Nevertheless, these CNVs should give useful information about the extent of copy number polymorphism in human populations.
The purpose of this article is to report the extent of CNVs for a few different types of sensory receptor genes and to relate it to the long-term change of copy numbers of the sensory receptor gene families. However, we first present the results for the entire set of annotated genes.
Results
CNVs of All Annotated Genes in Humans.
To have a general idea about the extent of CNVs in human populations, we identified all annotated genes with CNVs, which are included within each CNVR. We excluded the genes that are not included completely within the CNVRs from CNPGs, because a majority of the breakpoints of CNVRs are still ambiguous (19).
In this study, 3,144 of 22,218 human genes annotated in the Ensembl database (20) were identified as CNPGs. This number is similar to the previous estimate (2,908 genes) (13), which was obtained by using the RefSeq database (21). Our results indicate that 14.2% (3,144/22,218) of the human gene loci are polymorphic with respect to copy number. However, the number of polymorphic loci relative to the reference individual varied extensively among different sampled individuals. Fig. 1 shows the distribution of the number of loci in which the number of gene copies of a sampled individual is different from that of the reference individual. The ordinate stands for the number of individuals that showed a particular number of polymorphic loci relative to the reference individual. On average, a sampled individual had 277.2 loci different from the reference individual with respect to copy number. The maximum and minimum numbers of polymorphic loci were 574 and 108, respectively. These results indicate that an enormous number of CNPGs exist in human populations.
Fig. 1.
Distribution of the number of polymorphic loci in which a sampled individual shows a copy number different from that of the reference individual. All annotated loci were used. Mean and SD represent the mean and the standard deviation of the number of polymorphic loci, respectively.
Fig. 2 shows the distribution of relative copy number for all annotated genes among 270 individuals. The relative copy number represents the difference in copy number between the reference and a sampled individual. Here, it should be mentioned that when the copy number difference was identified between the reference and a sampled individual for a given gene (locus), the difference was assumed to be always one, because it was difficult to estimate the exact copy number difference from signal intensity data based on DNA hybridization (13). The distribution in Fig. 2 was approximately normal, and the mean of the distribution was 31.3 with a standard deviation (SD) of 54.4. This indicates that, on average, a randomly sampled individual contained a larger number of genes than the reference individual. To minimize the effect of the number of genes used on SD in the comparison of different gene families (see below), we computed the SD relative to the gene number in the standard human genome sequence (SDRG). (We could not compute the coefficient of variation, because the absolute copy number for each individual was not computable, as mentioned above.) This SDRG was 0.24% (54.4/22,218). When we computed the copy number difference for all possible pairs of individuals, the mean difference was 61.5, and in the most extreme case, one individual had 298 more genes than the other. The mean copy number difference relative to the gene number in the standard genome (MDRG) was 0.28% (61.5/22,218).
Fig. 2.
Distribution of relative copy number for all annotated genes. A curve represents the normal distribution fitted to the data. Mean and SD represent the mean and the standard deviation of gene copy number, respectively.
To evaluate the extent of CNVs within and among the three populations studied, we computed the relative copy number for each population (Fig. 3). In this analysis, we excluded offspring data, because the gene copy numbers of these individuals are correlated to those of their parental individuals. Each of the distributions roughly followed the normal distribution. On average, Africans had a larger number of gene copies than Asians or Europeans, and the differences in the mean numbers among the three populations were highly significant (P = 2.9 × 10−8 by F test). In addition, the African population showed the largest variation, which is consistent with the African origin hypothesis of modern humans (22, 23).
Fig. 3.
Distributions of relative copy number for all annotated genes in three human populations. N, sample size.
We then classified 3,144 CNPGs into the GO categories (15) to see the functional differences in CNVs. We found that the genes involved in sensory perception and immune response are significantly overrepresented [supporting information (SI) Table 4], as noted (13, 24).
CNVs of Sensory Receptor Genes in Humans.
Because the genes belonging to the olfactory receptor (OR) (16), taste receptor type 2 (T2R) (17), and vomeronasal (pheromone) receptor type 1 (V1R) (18) gene families are well characterized in the human genome, we investigated the CNVs for these genes in detail. [We did not analyze taste receptor type 1 and vomeronasal receptor type 2 genes, because the copy numbers of these gene families are very small in humans (6, 25).] For OR and T2R genes, we analyzed functional and nonfunctional genes separately. For V1R genes, only two of 117 genes have intact ORFs (18), but these genes are also suggested to be relics of an ongoing pseudogenization process (26). We therefore decided to regard all V1R genes as pseudogenes. Because the genomic locations of these genes were determined by using human genome assembly build 33 or 34 (16–18), whereas CNVR data were based on the build 35 (13), we reexamined the genomic locations of all genes using the build 35 (see Materials and Methods). After this reexamination, we obtained 796 OR (385 functional and 411 nonfunctional), 34 T2R (24 functional and 10 nonfunctional), and 113 V1R (all nonfunctional) genes.
Table 1 shows the numbers of polymorphic and monomorphic loci with respect to the copy number of sensory receptor genes. The proportion of polymorphic loci was 30% for functional OR genes and 35% for OR pseudogenes. Other genes also showed substantially higher proportions of polymorphic loci (40–80%) than the average for all annotated genes (14.2%). There was no significant difference in the proportion of copy number polymorphic loci between functional and nonfunctional OR or T2R genes (P > 0.1 by χ2 test).
Table 1.
Numbers of polymorphic and monomorphic loci with respect to the copy number for OR, T2R, and V1R genes
| Gene* | Polymorphic loci† | Monomorphic loci† | Total |
|---|---|---|---|
| OR (Funct) | 116 (30%) | 269 (70%) | 385 |
| OR (Pseudo) | 143 (35%) | 268 (65%) | 411 |
| T2R (Funct) | 12 (50%) | 12 (50%) | 24 |
| T2R (Pseudo) | 8 (80%) | 2 (20%) | 10 |
| V1R (Pseudo) | 45 (40%) | 68 (60%) | 113 |
*Funct, functional genes; Pseudo, pseudogenes.
†Numbers in parentheses represent the percentage of loci.
Fig. 4 shows the distributions of the relative copy numbers for OR, T2R, and V1R genes in all sampled individuals. The copy numbers for both functional and nonfunctional OR genes approximately followed the normal distribution, but those of T2R and V1R genes did not. When we computed the copy number difference for all possible pairs of individuals, the mean difference in functional OR genes was 10.9, and in the most extreme case one individual had 49 more genes than the other (Table 2). The other gene families also showed substantial CNVs among individuals. We found that SDRG is 1.9–15.8% [e.g., 2.5% (9.6/385) for functional OR genes], whereas MDRG is 2.1–10.0% [e.g., 2.8% (10.9/385) for functional OR genes] depending on the gene family. These values are much larger than the average for all annotated genes (0.24% and 0.28%, respectively). There was no significant difference in the variance of copy number between functional (91.8) and nonfunctional (97.3) OR genes (P = 0.63 by F test). However, because OR genes are frequently located in tandem in the human genome, the functional and nonfunctional OR genes are often included in the same CNVR. This may have the effect of equalizing the variances for functional and nonfunctional OR genes. We therefore excluded such CNVRs and reanalyzed the data. However, the variances for functional and nonfunctional OR genes were nearly the same (8.3 and 10.9, respectively).
Fig. 4.
Distributions of relative copy number of sensory receptor genes.
Table 2.
CNVs of sensory receptor genes in humans
| Gene* | Mean diff.† | Max. diff.‡ | SDRG§, % | MDRG¶, % |
|---|---|---|---|---|
| OR (Funct) | 10.9 | 49 | 2.5 | 2.8 |
| OR (Pseudo) | 11.3 | 52 | 2.4 | 2.7 |
| T2R (Funct) | 2.2 | 11 | 11.8 | 9.2 |
| T2R (Pseudo) | 1.0 | 8 | 15.8 | 10.0 |
| V1R (Pseudo) | 2.4 | 13 | 1.9 | 2.1 |
*Funct, functional genes; Pseudo, pseudogenes.
†Mean copy number difference between two individuals.
‡Maximum copy number difference between two individuals.
§SDRG, standard deviation relative to the gene copy number for a given gene family in the standard genome.
¶MDRG, mean copy number difference relative to the gene copy number for a given gene family in the standard genome.
We also analyzed the CNV of OR genes for each population separately (Fig. 5). All distributions were approximately normal. The mean relative numbers of functional OR genes of the three populations were significantly different from one another (P = 4.2 × 10−5 by F test), whereas the numbers of OR pseudogenes were not (P = 0.09). There was no significant difference between the variances of functional and nonfunctional OR genes in all populations.
Fig. 5.
Distributions of relative copy number of OR genes in three human populations.
CNVs of OR Genes Within and Between Populations or Species.
To examine the relationship between the copy number polymorphism within populations and the copy number divergence between populations, we computed the SD of copy number of OR genes in each population and the absolute values of the mean copy number difference between populations. Because we had three populations, the extent of polymorphism within populations was measured by the average of the SDs for the three populations, and the extent of divergence between populations was measured by the average of the absolute values of mean differences. The divergence relative to the polymorphism was measured by the ratio of the average of the mean differences to the average SD. We found that the extent of intrapopulational polymorphism is similar for functional genes and pseudogenes, but the extent of interpopulational divergence is greater for functional genes than for pseudogenes (Table 3).
Table 3.
CNVs of OR genes within and between populations or species
| Category* | Polymorphism† | Divergence‡ | Ratio§ |
|---|---|---|---|
| Within a human population vs. between human populations | |||
| Funct | 9.3 | 4.7 | 0.51 |
| Pseudo | 9.7 | 2.4 | 0.25 |
| Within humans vs. between humans and chimpanzees | |||
| Funct | 9.6 | 16 | 1.67 |
| Pseudo | 9.9 | 11 | 1.12 |
*Funct, functional genes; Pseudo, pseudogenes.
†Measured by the average of the SD of copy number within populations or species.
‡Measured by the average of the absolute value of the mean copy number differences between populations.
§Measured by the divergence relative to the polymorphism.
A similar analysis was conducted for CNVs for humans and chimpanzees. Because there are no comprehensive CNV data for chimpanzees, we assumed that the extent of polymorphism in chimpanzees is identical to that in the entire human population. We also assumed that the extent of the average species divergence is equal to the copy number difference of OR genes obtained by Y. Go and Y. Niimura (personal communication) from the standard human and chimpanzee genome sequences. There is another report about human and chimpanzee OR genes (27), but we did not use it, because it was based on early versions of the genome sequences. The ratios of the interspecific divergence to the intraspecific polymorphism were 1.67 and 1.12 for functional and nonfunctional genes, respectively, which are much higher than the corresponding ratios for human populations (Table 3). This result indicates that the copy number divergence has increased with evolutionary time.
Discussion
We have seen that human populations contain a large amount of CNVs of genes, and the MDRG is 0.28%, which is about three times higher than the proportion of single-nucleotide polymorphisms (SNPs) between two randomly chosen genomes (≈0.1%) (28–30). One may therefore argue that CNVs are more important than SNPs in generating phenotypic variation (13, 31), particularly if we note that most SNPs occur at silent nucleotide sites or noncoding regions (28).
It is interesting that the number of gene copies relative to the reference individual for all annotated genes approximately follows the normal distribution. This has occurred most likely because there are a large number of annotated genes that appear to have experienced duplication, deletion, and inactivation of genes, independently. It is well known that, if there are a large number of factors that contribute independently to a quantitative character, the character tends to show a normal distribution by the central limit theorem in probability theory.
We have also seen that the relative copy number of functional OR genes is approximately normally distributed in human populations. In this case, the number of gene loci subject to duplication and deletion is much smaller than in the case of all annotated genes. Yet the number of functional OR gene loci seems sufficiently large to generate the normal distribution of copy number. It is also possible that the normal distribution is generated by the birth–death process in probability theory (32). It is interesting to see that even the number of OR pseudogenes shows a normal distribution. Because pseudogenes are generated by various factors such as nonsense or frameshift mutations of functional genes, duplication of pseudogenes, etc., the normal distribution is likely to have been generated by the central limit theorem. Whatever the cause, OR pseudogenes are expected to evolve in a neutral fashion by genomic drift. Therefore, the normal distributions of functional and nonfunctional OR genes with similar variance suggest that the evolutionary change of functional OR genes is also largely controlled by random genomic drift, and the copy number change occurs in a more or less neutral fashion.
However, this does not mean that the number of copies of OR genes is unimportant for the ability of human olfaction. On the contrary, individuals with a larger number of OR genes may have a higher level of sensitivity to different odorants than those with a smaller number. Actually, it has been shown that polymorphisms in OR genes contribute to variability of odorant perception in humans (33). Nevertheless, olfaction is only one component of fitness for humans, and its contribution to total fitness may be minor in the presence of many other factors. For this reason, the number of OR genes may not be directly related to fitness.
Unlike OR genes, T2R and V1R genes did not follow the normal distribution. This is probably because the number of gene copies involved is small. In the case of T2R genes, however, the selective constraints are apparently relaxed in humans (34), and the copy number distributions of T2R genes is nearly the same for functional and nonfunctional genes. These results suggest that the CNV of functional T2R genes is also more or less neutral.
There is a common belief that the physiological requirement for a species is the major factor for determining the repertoire of a multigene family. For example, it has been hypothesized that the decrease in the number of OR genes in some primate species has occurred because these species acquired full trichromatic color vision, and this visual function has made olfaction less important (ref. 35, but see ref. 36 for correction). However, we have shown that the CNV of OR genes is large in humans, and the largest difference between two individuals in functional OR genes is 49, which is three times greater than the difference between the standard genomes of humans and chimpanzees. This result suggests that genomic drift plays an important role in the evolution of OR genes at least in humans.
Furthermore, the copy number change due to genomic drift may occasionally play important roles in phenotypic evolution (7). That is, when a new environmental niche is open for a species, and this niche requires a large number of genes, a group of individuals with large numbers of genes generated by genomic drift may move to this niche and eventually establish a new species. For example, terrestrial vertebrates have hundreds of class II OR genes, which are apparently for detecting airborne odorants (37). By contrast, teleost fishes have only a few genes that are orthologous to these class II genes. Obviously, these class II genes expanded enormously when terrestrial vertebrates evolved, and this expansion was probably aided by genomic drift.
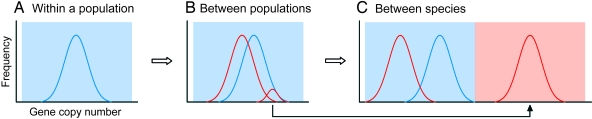
Fig. 6 shows a simple model of copy number evolution in sensory receptor genes. A natural population has substantial CNV (Fig. 6A). Here, the copy number is assumed to change mostly at random as long as the number is within the upper and lower boundaries determined by physiological requirements. When a population is separated into two populations, these populations may have different distributions of copy number largely because of genomic drift (Fig. 6B). This type of differentiation of populations may proceed even to generate different species (Fig. 6C). By contrast, a new species may be generated when a group of individuals having a larger number of genes by genomic drift (Fig. 6B) moves to a new niche, where a larger number of genes are needed (Fig. 6C).
Fig. 6.
Schematic diagram of copy number evolution in sensory receptor genes. (A) CNV within a population. (B) CNVs in two geographical populations. (C) CNVs in two species. Each color line represents the distributions of gene copy number for each species or population, whereas different color shades show different environmental conditions. A solid arrow indicates that a group of individuals with a larger number of genes than the average (B) moves to a new niche and establishes a new species (C).
It should be noted that a large extent of genomic drift is not confined to sensory receptor genes. It seems to have occurred also in the evolution of Ig genes (S. Das, M. Nozawa, J. Klein, and M. Nei, unpublished work). However, genomic drift appears to be less important in genes controlling the basic cellular process such as DNA repair and homologous recombination (38) or genes controlling the characters in the early stage of development. For example, the number of Hox genes concerned with the formation of body pattern of animals is ≈40 in most tetrapod species (39, 40). It should also be noted that even with sensory receptor genes, the extent of genomic drift seems to be smaller in Drosophila than in mammals (41, 42). Nevertheless, genomic drift is apparently an important evolutionary factor for generating random change of phenotypic characters. Because it can affect many different sets of genes, its significance in evolution may be much greater than random genetic drift of gene frequencies caused by finite population size. It would be important to study the nature and effect of this evolutionary factor in detail in the future.
Materials and Methods
Determination of CNVRs in Each Individual.
Redon et al. (13) identified CNVRs in 270 human individuals by using two experimental platforms [i.e., Whole Genome TilePath (WGTP) and Affymetrix GeneChip Human Mapping 500K early access arrays (500K EA)] and obtained two different sets of CNVR data for each individual. To make a single set of CNVR data for each individual, we merged these two CNVR data as follows. (i) All CNVRs identified in the European individual NA10851 in the 500K EA platform were eliminated, because NA10851 was used as the reference in the WGTP platform. (ii) When more than one CNVR overlapped in a sampled genome, these CNVRs were merged if all CNVRs were gain or loss events (compared with NA10851). (iii) If these CNVRs contained both gain and loss events, we excluded the CNVRs from the analysis, because it is quite unlikely that one individual has both duplication and deletion in a same genomic region. The genomic locations of CNVRs for each individual are shown in SI Table 5.
Determination of CNPGs and GO Analysis.
To determine CNPGs, we used the CNVR data obtained and the gene location data (Homo_sapiens.NCBI35.feb.pep.fa) annotated in the Ensembl database (20) (www.ensembl.org). If a gene is completely included within a CNVR in at least one of the sampled individuals, the gene was regarded as a CNPG. For the GO analysis, the GO categories assigned for the same type of genes were extracted from the gene annotation files (GenBank format) and the ontology file (gene_ontology_edit.obo) from the GO database (15) (www.geneontology.org) on March 20, 2007. Statistical analysis was conducted by using GO::TermFinder (43).
Reexamination of the Genomic Locations of OR, T2R, and V1R Genes.
We reexamined the genomic locations of OR, T2R, and V1R genes using the human genome assembly build 35 as follows. (i) We conducted a BLASTn (44) search against the genome sequence using each of the genes previously identified (16–18) as a query. (ii) For each query, we extracted the best hit sequence, which showed the lowest E-value. (iii) Aligning each pair of query and best hit sequences, we determined the genomic locations of the genes. (iv) If the best hit sequences were located on unassembled chromosomes, the sequences were eliminated from the analysis. (v) If more than one gene were mapped to the same genomic location, we used the gene showing the lowest E-value with query sequences and eliminated the other sequences. The genomic locations of these genes in build 35 are shown in SI Tables 6–8. The procedures for determining the CNPGs in these gene families were the same as above.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Yasuhiro Go and Yoshihito Niimura for providing unpublished data about the numbers of OR genes in humans and chimpanzees. We also thank Dimitra Chalkia, Saby Das, Hiroki Goto, Zhenguo Lin, Yoshihito Niimura, Nikos Nikolaidis, Helen Piontkivska, Alex Rooney, Shigeru Saito, Claire T. Saito, Yoko Satta, Yoshiyuki Suzuki, Shozo Yokoyama, and Jianzhi Zhang for comments on earlier versions of the manuscript. This work was supported by National Institutes of Health Grant GM020293 (to M. Nei).
Footnotes
The authors declare no conflict of interest.
See Commentary on page 20147.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709956104/DC1.
References
- 1.Rubin GM, Yandell MD, Wortman JR, Miklos GLG, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W, et al. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.International Human Genome Sequencing Consortium. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Ache BW, Young JM. Neuron. 2005;48:417–430. doi: 10.1016/j.neuron.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 4.Niimura Y, Nei M. J Hum Genet. 2006;51:505–517. doi: 10.1007/s10038-006-0391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niimura Y, Nei M. PLoS ONE. 2007;2:e708. doi: 10.1371/journal.pone.0000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi P, Zhang J. Genome Res. 2007;17:166–174. doi: 10.1101/gr.6040007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nei M. Proc Natl Acad Sci USA. 2007;104:12235–12242. doi: 10.1073/pnas.0703349104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 9.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Månér S, Massa H, Walker M, Chi M, et al. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 10.Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, et al. Nat Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 11.Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. Nat Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 12.Khaja R, Zhang J, MacDonald JR, He Y, Joseph-George AM, Wei J, Rafiq MA, Qian C, Shago M, Pantano L, et al. Nat Genet. 2006;38:1413–1418. doi: 10.1038/ng1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews D, Fiegler H, Shapero MH, Carson AR, Chen W, et al. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong KK, deLeeuw RJ, Dosanjh NS, Kimm LR, Cheng Z, Horsman DE, MacAulay C, Ng RT, Brown CJ, Eichler EE, et al. Am J Hum Genet. 2007;80:91–104. doi: 10.1086/510560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niimura Y, Nei M. Proc Natl Acad Sci USA. 2003;100:12235–12240. doi: 10.1073/pnas.1635157100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Go Y, Satta Y, Takenaka O, Takahata N. Genetics. 2005;170:313–326. doi: 10.1534/genetics.104.037523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young JM, Kambere M, Trask BJ, Lane RP. Genome Res. 2005;15:231–240. doi: 10.1101/gr.3339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korbel JO, Urban AE, Grubert F, Du J, Royce TE, Starr P, Zhong G, Emanuel BS, Weissman SM, Snyder M, et al. Proc Natl Acad Sci USA. 2007;104:10110–10115. doi: 10.1073/pnas.0703834104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubbard TJP, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pruitt KD, Tatusova T, Maglott DR. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cann RL, Stoneking M, Wilson AC. Nature. 1987;325:31–36. doi: 10.1038/325031a0. [DOI] [PubMed] [Google Scholar]
- 23.Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson AC. Science. 1991;253:1503–1507. doi: 10.1126/science.1840702. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen D-Q, Webber C, Ponting CP. PLoS Genet. 2006;2:e20. doi: 10.1371/journal.pgen.0020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao J, Schultz PG. Mamm Genome. 2003;14:291–301. doi: 10.1007/s00335-002-2233-0. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Webb DM. Proc Natl Acad Sci USA. 2003;100:8337–8341. doi: 10.1073/pnas.1331721100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilad Y, Man O, Glusman G. Genome Res. 2005;15:224–230. doi: 10.1101/gr.2846405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.International SNP Map Working Group. Nature. 2001;409:928–933. [Google Scholar]
- 29.Reich DE, Schaffner SF, Daly MJ, McVean G, Mullikin JC, Higgins JM, Richter DJ, Lander ES, Altshuler D. Nat Genet. 2002;32:135–142. doi: 10.1038/ng947. [DOI] [PubMed] [Google Scholar]
- 30.Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, et al. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beckmann JS, Estivill X, Antonarakis SE. Nat Rev Genet. 2007;8:639–646. doi: 10.1038/nrg2149. [DOI] [PubMed] [Google Scholar]
- 32.Feller W. An Introduction to Probability Theory and Its Applications. 2nd Ed. New York: Wiley; 1957. [Google Scholar]
- 33.Keller A, Zhuang H, Chi Q, Vosshall LB, Matsunami H. Nature. 2007;449:468–473. doi: 10.1038/nature06162. [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Thomas SD, Zhang J. Hum Mol Genet. 2004;13:2671–2678. doi: 10.1093/hmg/ddh289. [DOI] [PubMed] [Google Scholar]
- 35.Gilad Y, Wiebe V, Przeworski M, Lancet D, Pääbo S. PLoS Biol. 2004;2:e5. doi: 10.1371/journal.pbio.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilad Y, Wiebe V, Przeworski M, Lancet D, Pääbo S. PLoS Biol. 2007;5:e148. [Google Scholar]
- 37.Niimura Y, Nei M. Proc Natl Acad Sci USA. 2005;102:6039–6044. doi: 10.1073/pnas.0501922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin Z, Kong H, Nei M, Ma H. Proc Natl Acad Sci USA. 2006;103:10328–10333. doi: 10.1073/pnas.0604232103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoegg S, Meyer A. Trends Genet. 2005;21:421–424. doi: 10.1016/j.tig.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 40.Nam J, Nei M. Mol Biol Evol. 2005;22:2386–2394. doi: 10.1093/molbev/msi229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo S, Kim J. Mol Biol Evol. 2007;24:1198–1207. doi: 10.1093/molbev/msm038. [DOI] [PubMed] [Google Scholar]
- 42.Nozawa M, Nei M. Proc Natl Acad Sci USA. 2007;104:7122–7127. doi: 10.1073/pnas.0702133104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.