Abstract
A comprehensive phylogenetic framework is indispensable for investigating the evolution of genomic features in mammals as a whole, and particularly in humans. Using the ENCODE sequence data, we estimated mammalian neutral evolutionary rates and selective pressures acting on conserved coding and noncoding elements. We show that neutral evolutionary rates can be explained by the generation time (GT) hypothesis. Accordingly, primates (especially humans), having longer GTs than other mammals, display slower rates of neutral evolution. The evolution of constrained elements, particularly of nonsynonymous sites, is in agreement with the expectations of the nearly neutral theory of molecular evolution. We show that rates of nonsynonymous substitutions (dN) depend on the population size of a species. The results are robust to the exclusion of hypermutable CpG prone sites. The average rate of evolution in conserved noncoding sequences (CNCs) is 1.7 times higher than in nonsynonymous sites. Despite this, CNCs evolve at similar or even lower rates than nonsynonymous sites in the majority of basal branches of the eutherian tree. This observation could be the result of an overall gradual or, alternatively, lineage-specific relaxation of CNCs. The latter hypothesis was supported by the finding that 3 of the 20 longest CNCs displayed significant relaxation of individual branches. This observation may explain why the evolution of CNCs fits the expectations of the nearly neutral theory less well than the evolution of nonsynonymous sites.
Keywords: constrains, generation time, genome, population size
Different evolutionary forces shape the various classes of functional genomic elements. Whereas the majority of the genome evolves neutrally, functional elements undergo selection, either purifying selection (maintaining functions) or positive selection (favoring new functions) (1, 2).
Several diverse mechanisms that shape the evolution of a genome have recently been described. Of particular interest is the generation time (GT) hypothesis, which suggests that species with long GTs exhibit slower molecular clocks than species with short GTs because most germ-line mutations originate from errors in DNA replication (3–5). The GT hypothesis also explains the hominoid slowdown phenomenon, in which hominoids have been shown to evolve at a slower rate than Old World monkeys (cercopithecoids) (4–11). In contrast to the rest of a mammalian genome, CpG dinucleotides accumulate mutations independently from DNA replication. Substitutions in CpG sites occur relatively quickly and are constant over time because of methylation/deamination (12–14). Because CpG prone sites (followed by G or preceded by C) are hypermutable, they are often excluded from genomic comparisons (14–16).
The nearly neutral theory posits that all slightly deleterious and slightly beneficial mutations [with a selection coefficient (s) whose absolute value is less than the inverse of the effective population size (Ne)] behave as if they were neutral and may spread throughout the population because of random genetic drift (17). Because the effect of positive selection is negligible (i.e., most new mutations have s < 0), the nearly neutral theory deals mainly with slightly deleterious mutations. Given that the probability of fixation of slightly deleterious mutations depends on the effective population size, there is a class of these mutations that can be fixed in small populations because of random drift but are counter-selected, through purifying selection, from large populations. Thus, the nearly neutral theory predicts that the probability of fixation of slightly deleterious mutations and, consequently, the rate of evolution of constrained elements in small populations should be higher than in large populations (17, 18).
The prediction that species with small populations should have a higher probability of fixation of slightly deleterious mutations was corroborated in comparative studies of primates and rodents (10, 19, 20); mammals, birds, and drosophilids (21); island vs. continent-inhabiting populations of the same species (22); and large vs. small-bodied mammals (23).
Recent comparisons among several mammalian genomes have suggested that ≈5% of nucleotides have evolved under purifying selection and may therefore be functional (24–30). These constrained elements include protein-coding sequences (CDSs) and conserved noncoding sequences (CNCs). The latter group represents features such as transcriptional regulatory elements, matrix attachment regions, interchromosomal interactions, and other sequences of unknown function (31–36). Contrasting the modes of evolution between CDS and CNC genomic elements is important for understanding the difference in selection that acts on these candidate functional sequences. Previous attempts to determine the evolutionary patterns of these two classes of functional sequences used only a few species comparisons, such as the human, chimpanzee, mouse, and rat genomes (37–40).
In this study, we used a representative dataset of 44 different genomic regions selected by the ENCODE pilot project (36, 41, 42). By analyzing 1% of 18 mammalian genomes and their reconstructed ancestral states, we investigated how life-history traits drive molecular evolution on the genomic scale at synonymous sites (also known as silent sites), nonsynonymous sites, and CNCs.
Results and Discussion
We analyzed the pilot ENCODE sequence data of a representative set of mammalian species (36) to characterize and compare (i) neutral evolutionary rates [approximated by the number of synonymous substitutions per synonymous site (dS) (2)], (ii) evolutionary rates of constrained protein coding elements [represented by the number of nonsynonymous substitutions per nonsynonymous site (dN)], and (iii) evolutionary rates of conserved noncoding (CNC) genomic elements using a robust evolutionary framework (43). We created two alignments, one covering all coding sequences (205 kb, 218 genes) and the other containing all CNC elements longer than 15 bp (a total of 539 kb). Mammalian tree branch lengths were calculated separately for synonymous sites, nonsynonymous sites, and CNC sequences by using maximum likelihood methods [supporting information (SI) Fig. 5].
Neutral Evolutionary Rates Depend on GT.
We asked whether potentially neutral sites (represented here by synonymous sites) evolve according to the GT hypothesis. Trees based on substitutions at synonymous sites clearly show shorter branch lengths in primates, particularly in the human lineage. To test whether hominoids, and more specifically humans, display significantly lower synonymous substitution rates relative to other primates or non-primate mammals, we performed relative rate tests (44). We found that primates exhibit a significantly slower accumulation of substitutions at synonymous sites compared with all other mammalian lineages (P < 0.001) except for Xenarthra, which is represented by armadillo (P = 0.39) (SI Table 1). These results also clearly indicate that within primates, hominoids (chimpanzees and humans) have undergone a significant slowdown in the rate of silent substitutions compared with cercopithecoids (represented by macaques and baboons, P < 0.001). The trend of a slowdown in hominoids reaches a peak in humans, where dS is ≈2% smaller than in the chimpanzees (P < 0.05). Our results are compatible with the “hominoid slowdown” proposal, which may be related to the GT effect, because hominoids generally have longer GTs compared with other primates (7, 45–47).
Does the GT effect explain differences in other mammalian lineages as well? To address this question, we have calculated the ratios between the evolutionary rates of synonymous substitutions for 11 pairs of sister clades (dS1/dS2) and correlated them with the ratios of corresponding generation times (GT1/GT2) (SI Table 2). As expected by the GT hypothesis, our data show that species with longer GTs accumulate fewer synonymous substitutions over time. There is a robust linear regression between GT and dS (dS1/dS2 = 0.72533 − 0.01437 × GT1/GT2, R2 = 0.4787, P = 0.01833; Fig. 1) and a statistically significant inverse rank correlation (Kendall tau = −0.6363636, P = 0.002854).
Fig. 1.
Relationship between neutral evolutionary rates (dS) and GTs based on 11 sister lineage comparisons. For the sister taxa, plotted on the x axis are the values of the rates of the generation times (GT1/GT2), and plotted on the y axis are the values of the rates between branch lengths (dS1/dS2). Identification of each point is as in SI Table 2 (top-down numeration, so that 1 is the human/chimpanzee clade and 11 is the elephant/tenrec clade).
Because CpG prone sites are expected to evolve fast and may lead to underestimation of branch lengths due to saturation, we also created a CDS alignment depleted of CpG prone sites. This resulted in the exclusion of 39% (±0.8%) of the positions (SI Table 3). Kim et al. (14) concluded that CpG prone sites are not subjected to GT effects; therefore, by eliminating these sites, we expected an improved correlation between GT and dS. Indeed, with CpG prone sites excluded, we found a slightly better correlation between GT and dS [when the comparison was performed without the chimpanzee branch due to a very small dS and a large standard error (SE) (see Materials and Methods)]. After excluding the CpG prone sites, the length of the dS tree became 53% shorter.
As an additional approach, we used the phylogenetic independent contrasts (PIC) method, after which we obtained 16 statistically independent contrasts of GT and dS (see Materials and Methods). Using the contrasts, we constructed linear regressions through the origin, which were significant for complete sequences [n = 16: slope = −0.0094, P = 0.033] and marginally significant for sequences with eliminated CpG prone sites [n = 16: slope = −0.004382, P = 0.058; n = 15 (without contrast chimpanzee minus human): slope = −0.0047, P = 0.0565].
Evolutionary Rates of Constrained Elements Depend on the Effective Population Size.
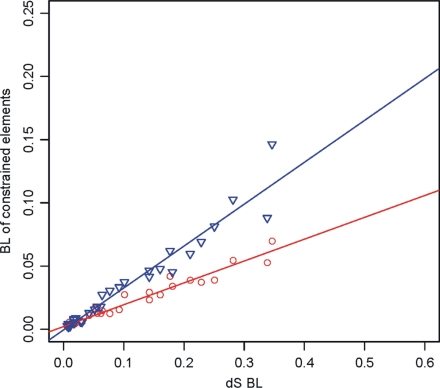
Because synonymous substitutions reflect the baseline rate at which genomes evolve, the substitution rate of constrained elements (nonsynonymous sites and CNCs) is expected to be proportionally lower depending on the intensity of negative selection acting on them. To assess the extent of selection at nonsynonymous and CNC sites, we estimated their evolutionary rates in a phylogenetic framework. For this purpose, we performed relative rate tests (SI Table 1) using fully resolved trees based on constrained elements. This analysis revealed a slowdown in evolutionary rates of nonsynonymous substitutions and CNCs in the genomes of primates (P < 0.001) similar to the effect detected for silent substitutions. Within primates, hominoids display lower evolutionary rates at nonsynonymous sites and CNC sequences than cercopithecoids (P < 0.01), whereas humans show the lowest absolute rates (P < 0.01 compared with chimpanzees). There is a strong correlation between the rate of neutral evolution (as approximated by synonymous changes) and the rate of constrained genomic elements (R2 > 0.94; Fig. 2). On average, nonsynonymous sites evolve 5.2 times more slowly than silent sites (red line in Fig. 2), whereas CNCs evolve 3.1 times more slowly than silent sites (blue line in Fig. 2).
Fig. 2.
Correlation of mammalian branch lengths between constrained elements [dN (red) and CNC (blue)] and neutral evolutionary rates (dS) categories.
The evolution of constrained elements may not be primarily driven by GT (18). Indeed, there is no significant correlation between GT and dN (linear regression: dN1/dN2 = 0.698402 − 0.005653 × GT1/GT2, R2 = 0.09065, P = 0.3683; Kendall rank correlation tau = −0.2568915, P = 0.1371), but there is a significant negative trend (although with a very small slope) when using the PIC method (slope = −0.00158, P = 0.0164 for original sequence; slope = −0.000747, P = 0.038 for sequences without CpG prone sites). This is most likely due to the fact that nonsynonymous sites undergo strong constraints and thus deviate from neutral evolution. The evolutionary rates of CNCs (CNCrate) are also not significantly correlated to GT by linear regression (CNCr1/CNCr2 = 0.741398 − 0.014054 × GT1/GT2, R2 = 0.2404, P = 0.1258). However, they do show a significant rank correlation (Kendall rank correlation tau = −0.4909091, P = 0.02027). The trend is also significant when using the PIC method (slope = −0.0034, P = 0.022 for original sequences; slope = −0.0028, P = 0.0154 for sequences without CpG prone sites).
The nearly neutral theory of molecular evolution predicts that mutations in nonsynonymous sites and CNC elements will be fixed predominantly in small populations where intensive genetic drift exists. To test this hypothesis, we first assessed the selective pressure acting on both types of constrained elements by dividing their evolutionary rates by dS for each branch of the tree, which displayed highly heterogeneous dN/dS (denoted as omega) and CNCrate/dS (CNCr/dS) ratios (SI Fig. 6). Calculation of CNCr/dS values is analogous to the approach of Lynch (48), in which the idea is extended to include both tRNA and rRNA genes, employing the ratio of the observed substitution rates in such DNA regions to the rate of synonymous substitutions in protein-coding genes.
The estimation of omega ratios for each branch of the tree (red bars in SI Fig. 6) reveals that despite the observed hominoid slowdown, this lineage has been accumulating more nonsynonymous substitutions per unit of silent substitutions than the vast majority of the mammalian lineages analyzed here. Of the 32 branches, 7 display a relaxation of negative selection with omega ratios clearly above the average (omega = 0.19), and these include the terminal branches of hominoids and cercopithecoids (omega > 0.28; SI Fig. 6). In contrast, branches leading to mouse, rat, and rabbit show the most extensive purifying selection acting on protein-coding regions, with omega values below 0.17. This result is consistent with previous observations, suggesting that purifying selection performs better in species with larger effective population sizes (Ne), such as rodents, as predicted by the nearly neutral theory of molecular evolution (38). Indeed, Ne was estimated to be ≈10,000 in humans and ≈30,000 in chimpanzees, whereas in mice the estimate is ≈85,000 (38, 49–51). Weak purifying selection acting on humans because of a small Ne has also been previously reported (52, 53).
We have extended the analysis of the relationship between selective pressure acting on nonsynonymous sites vs. Ne to the full set of available species where the Ne was approximated by GT (SI Table 3). Because GT and population size appear to be negatively correlated (54), we expected to observe a positive relationship between the probability of fixation of slightly deleterious mutations (omega of external branches of the tree) and GT. Estimates of omega per branch significantly correlated with GT (original alignment: omega = 0.177 + 0.010 × GT, R2 = 0.534, P < 0.0001; CpG prone minus alignment: omega = 0.0649 + 0.029 × GT, R2 = 0.605, P = 0.0001) (black line in Fig. 3) and exhibit significant rank correlation (original alignment: Kendall's tau = 0.6604, P < 0.0001; CpG prone minus alignment: Kendall's tau = 0.429, P = 0.006), corroborating the original prediction. If we account for phylogenetic non-independence of data and construct regression through the origin, which is based on independent contrasts, the trend is still significant (original alignment: slope = 0.0057, P = 0.0174; CpG prone minus alignment: slope = 0.0204, P = 0.0326). To view the relationship in more detail, we analyzed two new alignments (using the same approach) created either from conservative (blue line in Fig. 3) or less conservative sites (red line in Fig. 3) (as inferred from codeml with two discrete types of sites: 73.4% of conservative sites with average omega = 0.04 and 26.6% of less conservative sites with average omega = 0.66). There are significant regressions for conservative sites (linear regression: omega = 0.0391 + 0.004 × GT, R2 = 0.689, P < 0.0001; rank correlation: Kendall's tau = 0.5055, P = 0.002) as well as for nonconservative sites (linear regression: omega = 0.4396 + 0.0168 × GT, R2 = 0.543, P = 0.0005; rank correlation: Kendall's tau = 0.633, P = 0.0001). Both classes of sites demonstrate a positive trend, even if we account for phylogenetic non-independence (conservative sites: slope = 0.0074, P < 0.0001; less conservative sites: slope = 0.0537, P = 0.007). Because positive regressions between omega ratios and GTs are still significant for both conservative and less conservative sites, it seems likely that the accumulation of nearly neutral mutations drives evolution for all sites of the analyzed protein-coding genes.
Fig. 3.
Correlation of dN/dS (omega) ratios of 17 ENCODE species (terminal branches) to GTs (as an approximation of effective population size). Black line, dN/dS all codons; blue line, dN/dS conservative codons; red line, dN/dS less conservative codons.
Less conservative sites (red line in Fig. 3) accumulate slightly deleterious mutations more quickly when compared with conservative sites (blue line in Fig. 3); at these sites, both the intercept and slope values are low. The difference between intercepts and slopes for conservative and less conservative sites is most likely explained by the following: (i) a large fraction of mutations in less conservative sites are nearly neutral; and (ii) with the decrease of Ne (i.e., increase of GT), this fraction increases rapidly. In other words, relaxation of purifying selection with decreasing Ne is more pronounced for nonsynonymous sites, occurring in less conservative sites when compared with conservative ones.
GT Better Approximates Ne Compared with Body Mass.
We used GT as one approximation of Ne. However, it is well known that body mass (BM) also strongly influences population density and consequently population size (55). Thus, we can assess which life-history trait better relates to Ne (SI Table 3). As expected, log(BM) and GT exhibit a robust positive relationship (linear regression: log(BM) = 6.0537 + 0.4074 × GT, R2 = 0.410, P = 0.004; rank correlation: Kendall's tau = 0.660, P < 0.0001; regression based on independent contrasts: slope = 0.24955, P = 0.002). Next, we regressed omega ratios from BM. We observed that omega ratios for both conservative and less conservative sites significantly regress from BM according to the linear regression model (omega = 0.13878 + 0.0103 × log(BM), R2 = 0.228, P = 0.045), as well as to a rank correlation model (Kendall's tau = 0.385, P = 0.013). However, this trend almost fully disappears if we account for the effect of phylogeny (slope of <0.0001, P = 0.0976). Multiple linear regressions where GT and BM are two independent variables demonstrate that GT (not BM) is the only significant effector of regression for the omega ratio (P value for BM coefficient = 0.946187, P value for GT coefficient = 0.007). We can therefore assume that GT gives a better approximation of Ne when compared with BM.
CNC Evolution Deviates from the Expectations of the Nearly Neutral Theory.
We hypothesized that CNCs are evolving according to the same selective forces that act on protein coding sequences. It has been shown that the nearly neutral theory explains the evolution of CNCs based on human–chimpanzee and mouse–rat divergences (39). Surprisingly, our estimations of CNCr/dS per branch of the tree (blue bars in SI Fig. 6) do not always fit these expectations. Euprimates display the highest relaxation of purifying selection acting on CNCs (0.39), as presumed from the nearly neutral theory. However, notably Glires (rodents plus rabbits) show the second highest levels of such relaxation (0.36). They have higher CNCr/dS values than armadillo (0.35) or the cow plus dog lineage (0.25), and an equivalent value to elephant. The nearly neutral theory cannot explain these observations.
When we used GT to approximate Ne and extended the analysis to the whole set of mammalian species, we found a significant linear regression (CNCr/dS [original alignment] = 0.338 + 0.006 × GT, R2 = 0.265, P = 0.035; CNCr/dS [CpG prone minus alignment] = 0.583 + 0.033 × GT, R2 = 0.341, P = 0.014) that explains ≈1/3 of CNCr/dS variation but a nonsignificant rank correlation (Kendall tau for original alignment = 0.229, P = 0.100; Kendall tau for CpG prone minus alignment = 0.260, P = 0.073). When we accounted for phylogenetic non-independence of data and estimated the regression through the origin based on the independent contrasts, the trend disappeared (original alignment: P = 0.814; CpG prone minus alignment: P = 0.673). To assess other mechanisms driving the evolution of CNCs, we compared them to the rate of nonsynonymous substitutions.
Interestingly, the omega and CNCr/dS values differ significantly between branches (SI Fig. 6). On average, CNC sites evolve 1.7 times faster than nonsynonymous sites. In almost all recent taxa, the CNC branches are longer than the corresponding nonsynonymous substitution branches. However, we observed similar or higher levels of conservation in the CNC rates compared with dN in 7 of 10 mammalian deepest basal branches (those for cow–dog–bat, cow–dog, Primates, Euarchontoglires, Laurasiatheria, Boreoeutheria, and Exafroplacentalia). Assuming that the majority of genes maintain function and conservation through mammalian evolution, the observed differences with conservation patterns of CNC are probably due to a change of CNC mode of evolution.
Another possible explanation could be associated with the way in which CNCs were selected. The BinCons method (36) covers conserved elements all over mammalian taxa allowing for some variation in conservation. If the constraints acting on an individual CNC could relax in a lineage-specific manner, we would expect to find more instances in recent branches than in deep ones because early relaxing conserved elements would be less likely to fit the criteria of the BinCons method for selecting CNCs.
There are two possible explanations for why CNCs follow the expectations of the nearly neutral theory less well when compared with nonsynonymous sites. They may undergo (i) gradual relaxation along the phylogenetic tree or (ii) relaxation in individual CNCs in a lineage-specific manner (the turnover hypothesis). To test the gradual relaxation hypothesis, we contrasted the number of substitutions per million years in recent vs. deep branches of the tree. For this comparison, we used short branches to focus on a restricted time period. The set of recent branches included human, chimpanzee, hominoids, macaque, baboon, cercopithecoids, Catarrhini, mouse, and rat, whereas the set of deep branches was composed of Exafroplacentalia, Boreoeutheria, Euarchontoglires, Glires, Primates, and Laurasiatheria. No significant difference was detected between the two sets; consequently, this rules out the gradual relaxation hypothesis of evolutionary rates in CNCs (data not shown).
To test the turnover hypothesis, we analyzed the 20 longest individual CNCs from the ENCODE regions to detect whether CNCs are relaxed in particular lineages. We assessed branch lengths for those CNCs and compared them with the estimations derived from the concatenated CNC tree. From the set of 20 CNCs, 3 show one branch significantly longer than the branch in the concatenated CNC tree. Two branches are terminal (chimpanzee and shrew), and the third is the Eutheria branch (Fig. 4). These results support the turnover hypothesis rather than the gradual relaxation hypothesis in all CNCs. Thus, CNCs, conserved in all species, might undergo a relaxation or loss of constraints in a lineage-specific manner. This is consistent with the discovery of functional CNCs that are present in humans and missing in chimpanzees and macaques (35).
Fig. 4.
Three branches (Chimpanzee, Shrew, and Eutheria) are depicted on a concatenated CNC tree (first line) and on the 20 longest individual CNCs (in ENCODE regions). The branch length is plotted on the y axis (this reflects the number of substitutions), and the bars indicate the 95% confidence interval.
Contrary to protein coding genes, where nonfunctional genes (pseudogenes) are easily detected (for example by an omega ratio close to one), there is no obvious way to distinguish between functional and nonfunctional “dead” CNCs. To escape targeting CNCs with relaxed conservation pressures, they should be selected in a lineage-specific manner and not across distantly related species. Alternatively, the conservation pressure in each lineage can be assessed by comparing the branch lengths with our reference set of CNCs (SI Table 3).
The work presented here is a large-scale study investigating how life-history traits drive the evolution of genomic features. Our results provide a comprehensive picture validating and summarizing the proposed hypotheses with an unprecedented amount of genomic data covering the mammalian phyla. We conclude that (i) the neutral mutation rate depends on the GT; (ii) the evolutionary rates of constrained elements, especially nonsynonymous substitutions, depend partly, yet significantly, on the effective population size; and (iii) individual mammalian CNCs, as detected by ENCODE (38), might undergo a relaxation of constraints in a lineage-specific manner.
Materials and Methods
Alignments.
In the frame of the ENCODE project, we used the TBA alignments constructed by the Multi-species Sequence Alignment group (36). These alignments cover 30 Mb of human genome sequence taken from 44 different locations and include 18 mammalian species. From those alignments, we selected CDSs and CNCs. The CDS alignment was created by in-frame concatenation of the longest transcript per gene (same alignment as in ref. 43), whereas the CNC alignment includes all individual CNCs longer than 15 bp (SI Dataset 1). Because in the original ENCODE sequence alignment missing data and indels are marked by the same symbol, we treat both as missing data (“?”) here.
Sequence Analysis.
For inferring trees based on synonymous (dS) or nonsynonymous (dN) substitutions (SI Fig. 5), we used the codeml program of PAML (56) with a codon model specifying different transition/transversion rate ratios and different nucleotide frequencies for each codon position, without gene partitioning, and imposing the topology obtained in Nikolaev et al. (43) (run mode = 0). The tree based on CNCs was obtained by using baseml with the REV model and incorporating heterogeneous evolutionary rates over sites with 12 categories for the gamma distribution, and imposing the same topology. All sites with ambiguity characters or missing data were excluded (clean data = 1; 48,489 bp used for CDS of a total of 204,786 bp, and 106,945 bp used for CNCs of a total of 539,693 bp).
We calculated evolutionary rates of synonymous and nonsynonymous substitutions in CDSs for each branch of the mammalian tree, including the maximum number of sites. For that, we first reconstructed the ancestral sequence of each node of the tree using the codeml program of PAML (56) with the same codon model as mentioned above, and including all sites of the alignment (clean data = 0; 204,786 bp). Next, the dS and dN branch lengths and their SEs were obtained by calculating the substitution rates that occurred from node to node or node to tip by using the pairwise maximum likelihood estimation as implemented in the codeml program of PAML (run mode = −2). This allows for the inclusion of maximum sequence information yet excludes missing data (SI Table 3). The 95% confidence interval is 1.96 × SE. For all subsequent correlation analyses, we used the pairwise estimations of dN and dS (run mode = −2) and the batch estimation of CNC rates (clean data = 1).
The CpG prone sites were defined as nucleotide positions followed by G or preceded by C (14–16). New alignments for CDSs and CNCs excluding CpG prone sites were obtained by substituting these sites with the missing data characters (“?”). Branch length estimates for CDS and CNC datasets were then calculated following the same procedure as for the original datasets. In the pairwise analysis of the CDS data, CpG prone sites were also excluded from the ancestral sequences. The CpG prone sites comprise ≈39% in CDS and 30% in CNC alignments.
To assess the significance of the differences between the evolutionary rates of species or groups of species, we applied the relative rate test as implemented in the program RRTree (44).
Correlations between genomic mutation rates (dS, dN, and CNCrate) and GTs were drawn by calculating ratios between the branch lengths of sister clades (e.g., dS1/dS2) and of corresponding generation times (GT1/GT2). For each lineage, the average of the branch lengths from root to tip was used. We also performed the test with dS calculated on the CpG depleted CDS alignment. Because the number of retained dS substitutions in the branch leading to chimpanzee was extremely small (22 ± 5), comparison between human and chimpanzee resulted in a deviant point in the correlations. Therefore, the regression based on the alignments with deleted CpG prone sites was calculated without the chimpanzee branch. The lineages analyzed are presented in SI Table 2.
To eliminate the effect of phylogenetic inertia, which might compromise comparative species analyses, we used a standard method of PIC (57), which was implemented in the program COMPARE (58). The independent contrasts method starts with a set of comparative data and a phylogeny, and transforms these into a set of normally distributed, statistically independent and standardized “contrasts” that conform to the requirements of most parametric statistical tests. A linear regression of contrasts was fitted through the origin (59), with a prior check to ensure that the intercept of the full regression did not differ from zero. Thus, all relationships investigated in this work were checked by different methods, including standard linear regression, rank correlation, and PIC.
Supplementary Material
ACKNOWLEDGMENTS.
We thank the members of the ENCODE Multispecies Sequence Analysis group (leadership provided by G. M. Cooper and E.H.M.) for providing the sequence data, alignments, and CNCs; Drs. Samuel Deutsch, Shamil Sunyaev, and Henrik Kaessmann for help in the preparation of the article and valuable advice; and Vital-IT for computational support. This work was supported in part by the Intramural Research Program of the National Human Genome Research Institute at the National Institutes of Health, the European Union (Aneuploidy) (S.E.A.), and the Swiss National Science Foundation (S.E.A. and J.I.M.-B.). K.P. was supported by the Russian Academy of Sciences Molecular and Cellular Biology Program and Russian Foundation for Basic Research Grant 07-04-00521.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
**National Institutes of Health Intramural Sequencing Center Comparative Sequencing Program: Gerard G. Bouffard, Jacquelyn R. Idol, Valerie V. B. Maduro, and Robert W. Blakesley, Genome Technology Branch; Gerard G. Bouffard, Xiaobin Guan, Nancy F. Hansen, Baishali Maskeri, Jennifer C. McDowell, Morgan Park, Pamela J. Thomas, Alice C. Young, and Robert W. Blakesley, Intramural Sequencing Center.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705658104/DC1.
References
- 1.McDonald JH, Kreitman M. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 2.Nei M. Mol Biol Evol. 2005;22:2318–2342. doi: 10.1093/molbev/msi242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laird CD, McConaughy BL, McCarthy BJ. Nature. 1969;224:149–154. doi: 10.1038/224149a0. [DOI] [PubMed] [Google Scholar]
- 4.Li WH, Ellsworth DL, Krushkal J, Chang BH, Hewett-Emmett D. Mol Phylogenet Evol. 1996;5:182–187. doi: 10.1006/mpev.1996.0012. [DOI] [PubMed] [Google Scholar]
- 5.Yi S, Ellsworth DL, Li WH. Mol Biol Evol. 2002;19:2191–2198. doi: 10.1093/oxfordjournals.molbev.a004043. [DOI] [PubMed] [Google Scholar]
- 6.Bailey WJ, Fitch DH, Tagle DA, Czelusniak J, Slightom JL, Goodman M. Mol Biol Evol. 1991;8:155–184. doi: 10.1093/oxfordjournals.molbev.a040641. [DOI] [PubMed] [Google Scholar]
- 7.Goodman M. Hum Biol. 1961;33:131–162. [PubMed] [Google Scholar]
- 8.Goodman M, Barnabas J, Matsuda G, Moore GW. Nature. 1971;233:604–613. doi: 10.1038/233604a0. [DOI] [PubMed] [Google Scholar]
- 9.Koop BF, Goodman M, Xu P, Chan K, Slightom JL. Nature. 1986;319:234–238. doi: 10.1038/319234a0. [DOI] [PubMed] [Google Scholar]
- 10.Li WH, Tanimura M. Nature. 1987;326:93–96. doi: 10.1038/326093a0. [DOI] [PubMed] [Google Scholar]
- 11.Ellsworth DL, Hewett-Emmett D, Li WH. Mol Phylogenet Evol. 1993;2:315–321. doi: 10.1006/mpev.1993.1030. [DOI] [PubMed] [Google Scholar]
- 12.Bird AP. Nucleic Acids Res. 1980;8:1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duncan BK, Miller JH. Nature. 1980;287:560–561. doi: 10.1038/287560a0. [DOI] [PubMed] [Google Scholar]
- 14.Kim SH, Elango N, Warden C, Vigoda E, Yi SV. PLoS Genet. 2006;2:e163. doi: 10.1371/journal.pgen.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meunier J, Khelifi A, Navratil V, Duret L. Proc Natl Acad Sci USA. 2005;102:5471–5476. doi: 10.1073/pnas.0408986102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keightley PD, Lercher MJ, Eyre-Walker A. PLoS Biol. 2005;3:e42. doi: 10.1371/journal.pbio.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohta T. Theor Popul Biol. 1976;10:254–275. doi: 10.1016/0040-5809(76)90019-8. [DOI] [PubMed] [Google Scholar]
- 18.Ohta T, Gillespie JH. Theor Popul Biol. 1996;49:128–142. doi: 10.1006/tpbi.1996.0007. [DOI] [PubMed] [Google Scholar]
- 19.Ohta T. J Mol Evol. 1995;40:56–63. doi: 10.1007/BF00166595. [DOI] [PubMed] [Google Scholar]
- 20.Eyre-Walker A, Keightley PD, Smith NG, Gaffney D. Mol Biol Evol. 2002;19:2142–2149. doi: 10.1093/oxfordjournals.molbev.a004039. [DOI] [PubMed] [Google Scholar]
- 21.Keightley PD, Eyre-Walker A. Science. 2000;290:331–333. doi: 10.1126/science.290.5490.331. [DOI] [PubMed] [Google Scholar]
- 22.Woolfit M, Bromham L. Proc Biol Sci. 2005;272:2277–2282. doi: 10.1098/rspb.2005.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popadin K, Polishchuk LV, Mamirova L, Knorre D, Gunbin K. Proc Natl Acad Sci USA. 2007;104:13390–13395. doi: 10.1073/pnas.0701256104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dermitzakis ET, Reymond A, Lyle R, Scamuffa N, Ucla C, Deutsch S, Stevenson BJ, Flegel V, Bucher P, Jongeneel CV, Antonarakis SE. Nature. 2002;420:578–582. doi: 10.1038/nature01251. [DOI] [PubMed] [Google Scholar]
- 25.Dermitzakis ET, Reymond A, Scamuffa N, Ucla C, Kirkness E, Rossier C, Antonarakis SE. Science. 2003;302:1033–1035. doi: 10.1126/science.1087047. [DOI] [PubMed] [Google Scholar]
- 26.Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gregory SG, Sekhon M, Schein J, Zhao S, Osoegawa K, Scott CE, Evans RS, Burridge PW, Cox T, Fox V, et al. Nature. 2002;418:743–750. doi: 10.1038/nature00957. [DOI] [PubMed] [Google Scholar]
- 28.Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, et al. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 29.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Margulies EH, Blanchette M, Haussler D, Green ED. Genome Res. 2003;13:2507–2518. doi: 10.1101/gr.1602203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 32.Glazko GV, Koonin EV, Rogozin IB, Shabalina SA. Trends Genet. 2003;19:119–124. doi: 10.1016/S0168-9525(03)00016-7. [DOI] [PubMed] [Google Scholar]
- 33.Loots GG, Locksley RM, Blankespoor CM, Wang ZE, Miller W, Rubin EM, Frazer KA. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- 34.Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 35.Frazer KA, Tao H, Osoegawa K, de Jong PJ, Chen X, Doherty MF, Cox DR. Genome Res. 2004;14:367–372. doi: 10.1101/gr.1961204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Margulies EH, Cooper GM, Asimenos G, Thomas DJ, Dewey CN, Siepel A, Birney E, Keefe D, Schwartz AS, Hou M, et al. Genome Res. 2007;17:760–774. doi: 10.1101/gr.6034307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prabhakar S, Noonan JP, Pääbo S, Rubin EM. Science. 2006;314:786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- 38.Keightley PD, Kryukov GV, Sunyaev S, Halligan DL, Gaffney DJ. Genome Res. 2005;15:1373–1378. doi: 10.1101/gr.3942005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kryukov GV, Schmidt S, Sunyaev S. Hum Mol Genet. 2005;14:2221–2229. doi: 10.1093/hmg/ddi226. [DOI] [PubMed] [Google Scholar]
- 40.Xuan Z, Zhao F, Wang J, Chen G, Zhang MQ. Genome Biol. 2005;6:R72. doi: 10.1186/gb-2005-6-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.The ENCODE Project Consortium. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 42.The ENCODE Project Consortium. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nikolaev S, Montoya-Burgos JI, Margulies EH, Rougemont J, Nyffeler B, Antonarakis SE. PLoS Genet. 2007;3:e2. doi: 10.1371/journal.pgen.0030002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson-Rechavi M, Huchon D. Bioinformatics. 2000;16:296–297. doi: 10.1093/bioinformatics/16.3.296. [DOI] [PubMed] [Google Scholar]
- 45.Goodman M. Hum Biol. 1962;34:104–150. [PubMed] [Google Scholar]
- 46.Wu CI, Li WH. Proc Natl Acad Sci USA. 1985;82:1741–1745. doi: 10.1073/pnas.82.6.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elango N, Thomas JW, Yi SV. Proc Natl Acad Sci USA. 2006;103:1370–1375. doi: 10.1073/pnas.0510716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lynch M, Blanchard JL. Genetica. 1998;102/103:29–39. [PubMed] [Google Scholar]
- 49.Nachman MW. Genetics. 1997;147:1303–1316. doi: 10.1093/genetics/147.3.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu N, Jensen-Seaman MI, Chemnick L, Kidd JR, Deinard AS, Ryder O, Kidd KK, Li WH. Genetics. 2003;164:1511–1518. doi: 10.1093/genetics/164.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takahata N. Jpn J Genet. 1993;68:539–547. doi: 10.1266/jjg.68.539. [DOI] [PubMed] [Google Scholar]
- 52.Bustamante CD, Fledel-Alon A, Williamson S, Nielsen R, Hubisz MT, Glanowski S, Tanenbaum DM, White TJ, Sninsky JJ, Hernandez RD, et al. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- 53.Williamson SH, Hernandez R, Fledel-Alon A, Zhu L, Nielsen R, Bustamante CD. Proc Natl Acad Sci USA. 2005;102:7882–7887. doi: 10.1073/pnas.0502300102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chao L, Carr D. Evolution (Lawrence, Kans) 1993;47:688–690. doi: 10.1111/j.1558-5646.1993.tb02124.x. [DOI] [PubMed] [Google Scholar]
- 55.Damuth HD, Jr, Diamond AB, Rappoport AS, Renner JW. Am J Neuroradiol. 1983;4:1239–1242. [PMC free article] [PubMed] [Google Scholar]
- 56.Yang Z. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 57.Felsenstein J. Am Nat. 1985:1–15. [Google Scholar]
- 58.Martins E. COMPARE. Indiana Univ, Bloomington: Dept of Biology; 2004. Version 4.6b. [Google Scholar]
- 59.Garland T, Harvey PH, Ives AR. Syst Biol. 1992;41:18–32. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.