Abstract
SgrS is a 227-nt small RNA that is expressed in Escherichia coli during glucose-phosphate stress, a condition associated with intracellular accumulation of glucose-6-phosphate caused by disruption of glycolytic flux. Under stress conditions, SgrS negatively regulates translation and stability of the ptsG mRNA, encoding the major glucose transporter, by means of a base pairing-dependent mechanism requiring the RNA chaperone Hfq. SgrS activity mitigates the effects of glucose-phosphate stress, and the present study has elucidated a function of SgrS that is proposed to contribute to the stress response. The 5′ end of SgrS, upstream of the nucleotides involved in base pairing with the ptsG mRNA, contains a 43-aa ORF, sgrT, that is conserved in most species that contain SgrS-like small RNAs. The sgrT gene is translated in E. coli under conditions of glucose-phosphate stress. Analysis of alleles that separate the base pairing function of SgrS from the sgrT coding sequence revealed that either of these functions alone are sufficient for previously characterized SgrS phenotypes. SgrS-dependent down-regulation of ptsG mRNA stability does not require SgrT and SgrT by itself has no effect on ptsG mRNA stability. Cells expressing sgrT alone had a defect in glucose uptake even though they had nearly wild-type levels of PtsG (IICBGlc). Together, these data suggest that SgrS represents a previously unrecognized paradigm for small RNA (sRNA) regulators as a bifunctional RNA that encodes physiologically redundant but mechanistically distinct functions contributing to the same stress response.
Keywords: riboregulation, RNA stability, small proteins, phosphoenolpyruvate phosphotransferase system, glycolytic flux
Central metabolism is controlled by a complex regulatory network at many levels, including transcription, translation, and allosteric control of enzymes. Although it has been the subject of decades of research, knowledge of the mechanisms controlling glycolytic flux are still incomplete, even in the well studied model organism Escherichia coli. Numerous studies have noted that bacterial strains with an impaired capacity to metabolize phosphorylated sugars (including some of the substrates of glycolysis) often show strong phenotypes of growth inhibition or in some cases cell lysis (1–3). However, until recently, little was known about the mechanisms used by bacterial cells to deal with metabolic stress associated with intracellular phosphosugar accumulation. One form of phosphosugar stress occurs in pgi (phosphoglucose isomerase) mutant strains where stress is associated with accumulation of glucose-6-phosphate (G6P) when cells are exposed to glucose (4–6). An analogous condition occurs in wild-type strains exposed to the nonmetabolizable glucose analog α-methyl glucoside (αMG), resulting in accumulation of αMG-6-phosphate. In both situations, cell growth is inhibited and there is a specific destabilization of the ptsG mRNA, which encodes the major glucose transporter of the phosphoenolpyruvate phosphotransferase system (PTS) in E. coli (PtsG, IICBGlc) (4, 5). This posttranscriptional regulation of the ptsG mRNA under conditions of G6P accumulation (glucose-phosphate stress) suggested the existence of at least one specific regulatory response to deal with such stresses.
We discovered a small RNA regulator, SgrS, that is induced under glucose-phosphate stress conditions and is responsible for destabilization of ptsG mRNA (6). Expression of SgrS during glucose-phosphate stress is clearly important for the adaptation to stress because sgrS mutant strains are strongly inhibited compared with wild-type strains under these conditions (6). The negative regulation of translation and stability of ptsG mRNA by SgrS stops synthesis of glucose transport proteins (7) and is hypothesized to limit further accumulation of G6P or αMG6P. SgrS is encoded divergently from sgrR, which encodes the transcriptional activator required for SgrS synthesis (6). SgrS is 227 nt in length and was originally identified on the basis of its binding to the RNA chaperone Hfq (8). Previously characterized small RNA (sRNA) regulators that require Hfq for function regulate mRNA targets via sRNA:mRNA base pairing interactions, a mechanism referred to as riboregulation. Hfq binding stabilizes most sRNAs and in some cases remodels secondary structure to facilitate base pairing (9–11) and increase the rate of sRNA:mRNA association (12). SgrS activity on ptsG mRNA requires an Hfq-mediated base pairing interaction between sequences at the 3′ end of SgrS (Fig. 1) and sequences in the 5′ untranslated region of ptsG mRNA. Although many sRNAs have multiple mRNA targets, our unpublished studies to identify other putative SgrS targets revealed only two additional candidates that might be down-regulated by SgrS at the level of mRNA stability. Again, sequences at the 3′ end of SgrS were predicted to base pair with these other targets, leaving open the question of the role of the SgrS 5′ end.
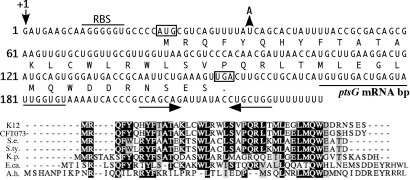
Fig. 1.
The SgrS sRNA contains a conserved ORF, sgrT. (Upper) The SgrS sequence is shown with the translated product of sgrT below the nucleotide sequence. The putative ribosome binding site for sgrT is indicated by the horizontal bar labeled “RBS.” The start and stop codons for sgrT are boxed. The fifth codon of sgrT was mutated to a “TAA” by a single base pair substitution indicated by the arrow and “A” (described in the text and in Fig. 2). The sequences involved in base pairing with the ptsG mRNA (7) are indicated by the horizontal line (labeled “ptsG mRNA bp”) below the nucleotide sequences. The inverted repeat that forms the terminator at the 3′ end of SgrS is indicated by horizontal arrows below the nucleotide sequence. (Lower) The SgrT amino acid sequences from E. coli K12 (K12) and other bacterial species were aligned with ClustalW. Matches to the consensus are shaded. CFT073, E. coli CFT073; S.e., Salmonella enterica paratyphi ATCC9150; S.ty., Salmonella enterica serovar Typhi Ty2; K.p., Klebsiella pneumoniae KP32; E.ca., Erwinia carotovora atroseptica SCRI1043; A.h., Aeromonas hydrophila ATCC7966.
Although there are several mechanisms of regulation by sRNAs, most regulatory sRNAs in bacteria are ≈100 nt long, act by base pairing with mRNA targets, and are not predicted to encode protein products. One exception is the 500-nt RNA III in Staphylococcus aureus that functions by base pairing with several mRNA targets (13–15) and also encodes the small protein delta hemolysin. RNA III is somewhat unique in that it appears that the base pairing-dependent regulation performed by RNA III does not require Hfq (16). In this study, we report SgrS as an example of an Hfq-dependent sRNA regulator that also encodes a functional protein product. This opens up the possibility that there are other such bifunctional sRNAs that have not yet been discovered and expands the mechanistic repertoire of these versatile regulators.
Results
The Small Regulatory RNA SgrS Contains a Conserved ORF, sgrT.
We have designated the ORF within the SgrS RNA sequence sgrT. This ORF is located at the 5′ end of SgrS and encodes a 43-aa polypeptide (Fig. 1); sequences that base pair with the ptsG mRNA are downstream from the stop codon of sgrT. SgrS-like sRNAs are fairly well conserved and can be found in the same genomic location in several bacterial species (6). Examination of SgrS orthologs from uropathogenic E. coli CFT073, Salmonella species, Klebsiella pneumoniae, and Erwinia carotovora revealed sgrT sequences that are quite well conserved with that of E. coli K12 (Fig. 1). Another putative sgrT ortholog was identified in Aeromonas hydrophila, a Gram-negative organism more distantly related to the aforementioned enteric species (Fig. 1). Interestingly, E. coli 0157:H7 and Yersinia pestis have SgrS orthologs; however, these RNAs do not appear to contain sgrT. The published sequence for E. coli 0157:H7 indicates that the sgrT start codon contains a substitution that alters the sequence to “ATT.” The SgrS ortholog in Y. pestis is well conserved at the 3′ end (where base pairing sequences reside) but is truncated at the 5′ end relative to the other SgrS orthologs and does not appear to contain sgrT. Despite these exceptions, sgrT is conserved in the majority of organisms with an sgrS ortholog, suggesting that it encodes a functionally important protein.
Base Pairing and SgrT Functions both Play a Role in the Glucose-Phosphate Stress Response.
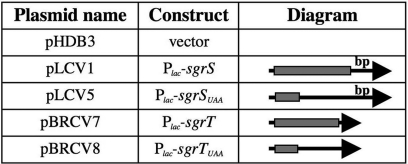
Because the sequences involved in base pairing with ptsG mRNA are downstream from sgrT, several constructs were created (Fig. 2) to individually analyze these two putative functions of SgrS. All of the sgrS and sgrT derivatives were cloned on plasmids and were expressed under the control of a heterologous promoter (Plac) that is IPTG-inducible. Wild-type SgrS possesses both base pairing and sgrT sequences [pPlac-sgrS or pLCV1; supporting information (SI) Table 1 and Fig. 2]. [This construct was previously shown to complement a chromosomal ΔsgrS::kan mutation (6).] A single-base substitution in the fifth codon of sgrT created a “UAA” stop codon, resulting in plasmid pPlac-sgrSUAA (pLCV5; SI Table 1 and Figs. 1 and 2). SgrSUAA should be able to perform the riboregulatory function of SgrS but not produce SgrT protein. To examine SgrT function in the absence of the base pairing sequences of SgrS, sgrT was cloned with a heterologous ribosome binding site (RBS), resulting in plasmid pPlac-sgrT (pBRCV7; SI Table 1 and Fig. 2). As a negative control, a “UAA” stop codon was placed at the fifth codon of sgrT, resulting in plasmid pPlac-sgrTUAA (pBRCV8; SI Table 1 and Fig. 2). This construct lacks the base pairing sequences and does not produce SgrT.
Fig. 2.
Construction of alleles that separate base pairing and SgrT functions. Plasmids are described in more detail in Methods and SI Table 1. All constructs are under the control of the Plac promoter. The SgrS molecule is depicted as an arrow, where the arrowhead is the 3′ end of SgrS. The location of sgrT is represented by the shaded rectangle at the 5′ end, and the location of the base pairing sequences at the 3′ end is indicated by the label “bp.” sgrSUAA contains a stop codon that truncates the sgrT ORF. The sgrT construct lacks the base pairing region. The sgrTUAA construct lacks the base pairing region and also contains a truncated sgrT ORF.
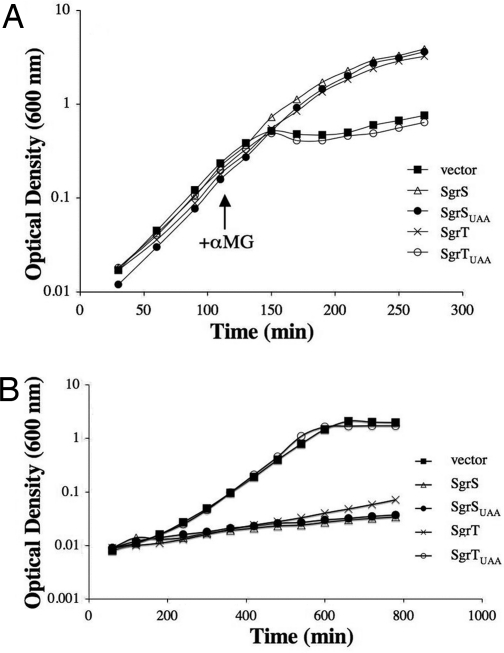
We showed previously that cells with wild-type SgrS have two prominent phenotypes related to glucose utilization and the glucose-phosphate stress response. SgrS activity is necessary for full recovery from glucose-phosphate stress caused by exposure of cells to αMG (6). Overexpression of SgrS also strongly inhibits growth when glucose is the sole carbon source (6). We originally hypothesized that both of these phenotypes were due to the base pairing function of SgrS and its activity on mRNA targets. Upon discovering sgrT within the SgrS sRNA, we set out to test whether SgrT also participates in glucose metabolism or stress physiology. A ΔsgrS::kan, lacIq+ host (where expression of alleles is repressed in the absence of IPTG) carrying plasmids described in Fig. 2 was analyzed for αMG stress recovery and growth on glucose. For αMG stress recovery, cells were grown with inducer (IPTG) to early logarithmic phase, and αMG was added to induce stress. As shown previously (6), cells lacking sgrS (vector; Fig. 3A) were strongly inhibited and failed to recover from stress, whereas cells carrying pPlac-sgrS (SgrS; Fig. 3A) recovered well. Cells expressing the sgrSUAA allele were able to recover from stress as well as cells expressing wild-type sgrS (Fig. 3A). This result suggested that sgrT is not absolutely essential for stress recovery under these conditions when the SgrS base pairing function is intact. Expression of the sgrT allele also rescued cell growth (Fig. 3A). The sgrTUAA allele failed to rescue growth (Fig. 3A), confirming that the SgrT phenotype was attributable to a functional SgrT polypeptide and not to nucleotide sequences within the 5′ region of SgrS. The same loss of rescue was observed with another sgrT allele where the ATG start codon was changed to a TAA stop codon (SI Fig. 7). Together, these results provided strong genetic evidence that sgrT encodes a functional protein that can participate in the glucose-phosphate stress response. The fact that alleles that possessed either base pairing (sgrSUAA) or SgrT (sgrT) functions could rescue cells from stress suggests that these two properties of the SgrS sRNA may be physiologically redundant.
Fig. 3.
Effect of SgrS base pairing and SgrT on recovery from glucose-phosphate stress and growth in glucose minimal medium. (A) A ΔsgrS::kan, lacIq+ host strain (CV104) carrying plasmids (pHDB3, pLCV1, pLCV5, pBRCV7, and pBRCV8) with alleles described in Fig. 2 was grown in LB with ampicillin and IPTG. Cells were stressed by addition of 0.5% αMG at early log phase. The data shown are representative of at least three independent experiments. (B) The strains described in A were grown in minimal A medium with glucose in the presence of ampicillin and IPTG. The results shown are representative of at least three independent experiments.
These same alleles were tested for the other known SgrS phenotype: growth inhibition when glucose is the sole carbon source. Cells expressing the constructs shown in Fig. 2 were grown in minimal glucose medium. Vector-containing cells grew to a high final cell density (Fig. 3B). As expected, cells overexpressing wild-type SgrS were strongly inhibited and failed to grow significantly (Fig. 3B, compare vector and SgrS). Interestingly, both SgrSUAA and SgrT inhibited cell growth on glucose to nearly the same degree as wild-type SgrS (Fig. 3B). Cells expressing SgrTUAA were not inhibited and grew as well as vector-containing cells. These results suggested that either riboregulation or SgrT function is sufficient for this phenotype.
A previous study showed that overexpression of SgrS caused growth inhibition primarily on glucose and slightly on mannose but not on other carbon sources tested (6). The specificity of SgrT-mediated growth inhibition was tested by streaking SgrT-overproducing cells on minimal media with one of a number of different carbon sources. On plates, SgrT-dependent growth inhibition was most striking when glucose was the sole carbon source. SgrT-overproducing cells were slightly inhibited for growth on mannose and N-acetylglucosamine but not detectably inhibited on mannitol, fructose, or casamino acids (SI Fig. 8). One of the predicted targets for SgrS riboregulation that emerged from microarray studies (C.K.V. and S. Gottesman, unpublished data) is the polycistronic manXYZ message. These genes encode a PTS transporter of relatively broad sugar specificity, including mannose and N-acetylglucosamine. The fact that SgrT overproduction inhibits growth on mannose and N-acetylglucosamine is consistent with other data (Fig. 3) that suggest that SgrS riboregulation and SgrT functions are somehow redundant.
To obtain biochemical evidence that sgrT phenotypes were associated with a specific protein product, an epitope-tagged SgrT protein was constructed and its production monitored by Western blot analysis. A sequence specifying three tandem FLAG epitopes (3XFLAG) was inserted at the 3′ end of sgrT in the context of pPlac-sgrT. The Plac-sgrT3XFLAG allele rescued cells from αMG stress and caused growth inhibition on glucose (data not shown), indicating that the epitope at the C terminus of SgrT did not significantly interfere with the function of the protein. A Western blot revealed that these cells produced a protein that migrated at ≈8 kDa, as predicted for SgrT-3XFLAG (SI Fig. 9). When the UAA mutation was introduced in the SgrT-3XFLAG construct, the protein was no longer made (SI Fig. 9).
sgrT Is Translated Under Glucose-Phosphate Stress Conditions.
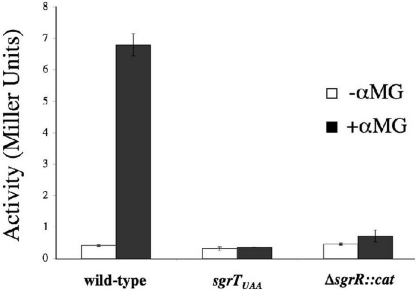
In the experiments described above, sgrT was ectopically expressed from a foreign promoter. To determine whether sgrT is translated under glucose-phosphate stress conditions in its native context, an sgrT′-′lacZ translational fusion was constructed. This fusion was placed in the natural chromosomal locus and joins sgrT coding sequence at the 38th codon with ′lacZ. The fusion was constructed in three strain backgrounds: wild-type, sgrTUAA, and ΔsgrR::cat. Because insertion of the fusion at the native locus effectively renders the fusion strains sgrS null mutants, a low concentration of αMG was used to avoid strong growth inhibitory effects. Strains were grown in rich (TB) medium and exposed to αMG in mid-log phase. β-galactosidase assays were performed on samples collected at several times after the addition of αMG. The basal level of activity in all strains in the absence of stress was very low (Fig. 4), but addition of αMG induced sgrT′-′lacZ activity in the wild-type background by ≈14-fold (Fig. 4). The data shown are from 3 h after induction; activity continued to increase as long as cells were cultured in the presence of αMG, reaching a level 25-fold greater than in uninduced cells after overnight culture (data not shown). When a stop codon was inserted upstream of the fusion junction (sgrTUAA), αMG-inducible activation of the fusion was abrogated. This indicates that sgrT is indeed translated under glucose-phosphate stress conditions in its native context. In the ΔsgrR::cat background, activation of the fusion was also eliminated (Fig. 4). This result is not surprising because we have shown that synthesis of the sRNA SgrS (which encodes sgrT) under stress conditions depends on the transcription factor SgrR (6, 17).
Fig. 4.
Translation of sgrT is activated under stress conditions in an SgrR-dependent manner. All strains carry a translational sgrT′-′lacZ fusion at the native sgrT locus. Strains BH300 (wild-type), BH301 (sgrTUAA), and BH302 (ΔsgrR::cat) were grown in rich medium to mid-log phase and split, and half of each culture was exposed to 0.005% αMG. β-galactosidase activity was measured at several time points thereafter. The data displayed are the average of three experimental trials at 3 h after exposure.
SgrT Does Not Participate in Posttranscriptional Regulation of the ptsG mRNA.
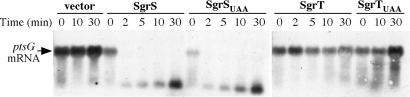
The hypothesis that SgrT may be redundant or cooperate with the base pairing function of SgrS in inhibiting translation and causing degradation of the ptsG mRNA was tested. In previous work, we showed that upon glucose phosphate stress, levels of SgrS increase, whereas levels of the ptsG mRNA diminish rapidly in an SgrS-dependent manner (6). Aiba and coworkers (4) showed that declining levels of ptsG message were due to the RNase E-dependent destabilization of the ptsG mRNA. To determine whether SgrT is involved in destabilizing the ptsG mRNA, the alleles shown in Fig. 2 were expressed in a lacIq+, sgrS mutant host, and the levels of ptsG mRNA were monitored at different time points after expression. As observed previously (6), in the absence of sgrS, the levels of ptsG mRNA remain steady, whereas when SgrS is expressed, the ptsG mRNA disappears (Fig. 5) because of RNase E-dependent degradation (4). The SgrSUAA variant also caused disappearance of the ptsG mRNA, suggesting that the base pairing function of SgrS does not require SgrT and is sufficient for posttranscriptional regulation of the ptsG mRNA. Northern blot analysis probing for SgrS and SgrSUAA showed that these two molecules accumulate after induction to approximately the same levels (SI Fig. 10), suggesting that the UAA mutation does not have a deleterious effect on the stability of SgrSUAA. When SgrT alone was expressed, levels of the ptsG mRNA did not change significantly (Fig. 5). Likewise, the negative control SgrTUAA did not alter levels of the ptsG mRNA (Fig. 5). These results indicate that the role of SgrT is independent of the base pairing function of SgrS and that SgrT acts at a different level in the glucose-phosphate stress response.
Fig. 5.
SgrT does not affect levels of ptsG mRNA. The ΔsgrS::kan, lacIq+ strain CV104 carrying constructs depicted in Fig. 2 was grown to mid-log phase in LB with ampicillin. Northern blot analysis was performed on total RNA extracts harvested at times indicated after cells were exposed to IPTG to induce expression of sgrS and sgrT constructs. The blot was probed for ptsG mRNA (indicated at the left).
Base Pairing and SgrT Functions both Inhibit Glucose Uptake but by Different Mechanisms.
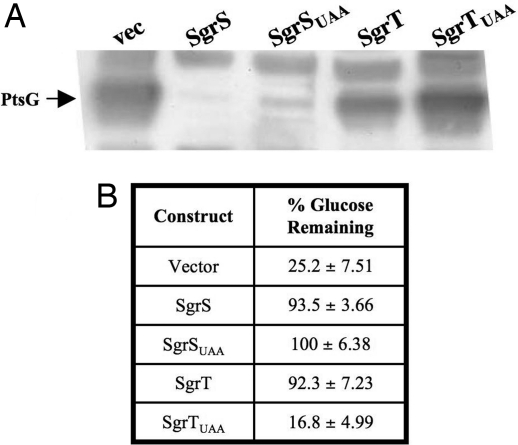
Wild-type cells recover from glucose-phosphate stress in a relatively short period and continue growing. Our current model for SgrS riboregulation of the ptsG message provides a mechanism for the cell to reduce influx of sugar phosphates indirectly by reducing the production of new sugar transport proteins. SgrT might contribute to recovery by a different mechanism; for example, by reducing the levels or activity of preexisting PtsG proteins. To examine the role of SgrS base pairing and SgrT on steady state levels of PtsG protein, the alleles described in Fig. 2 were expressed in a lacIq+, sgrS mutant background, and Western blot analysis was used to detect PtsG (IICBGlc). Levels of the PtsG protein were significantly lower in cells expressing SgrS and SgrSUAA compared with vector control cells (Fig. 6A). This result is consistent with the idea that SgrS molecules that can perform base pairing-dependent down-regulation of the ptsG mRNA stop new synthesis of PtsG protein, and preexisting PtsG is diluted as the cells continue to grow. PtsG levels in cells expressing SgrSUAA (containing the premature stop codon in sgrT) were approximately equivalent to levels in cells expressing the wild-type SgrS. This result again suggests that SgrT is not required and does not significantly contribute to the riboregulatory function of SgrS. Cells expressing SgrT alone had high levels of PtsG protein, as did cells expressing the negative control SgrTUAA (Fig. 6A). This result strongly suggests that SgrT does not function by promoting degradation of preexisting PtsG protein and led to the hypothesis that SgrT inhibits the transport activity of PtsG.
Fig. 6.
SgrS base pairing and SgrT functions individually block glucose uptake by different mechanisms. (A) Strains carrying plasmid constructs (as in Fig. 2) were grown in MOPS defined medium with glucose and amino acids with IPTG. Western blot analysis was performed on total protein extracts from samples harvested after 5 h of growth. The blot was probed for PtsG by using an αIIBGlc antibody. The position of the PtsG (IICBGlc) protein is indicated at left. Bands above and below the PtsG band are cross-reacting proteins and served as loading controls. (B) Strains are as described in A. Cells were harvested after 5 h of growth, and the amount of glucose remaining in the medium was measured. The numbers reported represent the amount of glucose remaining at 5 h divided by the amount of glucose in media before inoculation (% glucose remaining). The average of three independent experiments is reported.
The data above show that SgrT does not affect levels of ptsG mRNA or PtsG protein during stress. If SgrT acts at a posttranslational level on PtsG to inhibit its activity, this could account for the stress rescue and glucose growth inhibition phenotypes (Fig. 3). Alternatively, SgrT might interfere with metabolism of glucose at a step subsequent to transport. To test the hypothesis that SgrT interferes with glucose transport, the lacIq+, sgrS mutant strain with induced sgrS and sgrT alleles (as shown in Fig. 2) was grown in defined medium (MOPS) with glucose and amino acids. The growth rate of cells expressing SgrS, SgrSUAA, and SgrT was reduced compared with vector control and SgrTUAA cells (SI Fig. 11). (Growth of the former strains was not completely inhibited, as in Fig. 3B, because the medium used in this experiment contains amino acids that can be used as a carbon source.) The amount of glucose remaining in the supernatant was measured and normalized to the amount in the medium before cell inoculation (set at 100%). Cells carrying the vector control and negative control sgrTUAA plasmids used a significant portion of the glucose present; only ≈20% of the initial amount remained (Fig. 6B). In contrast, culture supernatants from cells expressing sgrS, sgrSUAA, or sgrT contained >90% of the initial amount of glucose (Fig. 6B), indicating that expression of each of these alleles prevented cells from taking up glucose. Together, these results support the model that the mechanism for inhibition of glucose uptake in cells expressing base pairing-proficient SgrS molecules is strong reduction of the amount of glucose transport protein. In contrast, SgrT-mediated inhibition of glucose uptake appears to occur at the level of PtsG activity.
SgrT Overexpression Prevents Inducer Exclusion.
The glucose-specific PTS enzyme IIAGlc phosphorylates IICBGlc (PtsG) and also has important regulatory functions in carbon catabolite repression (18). The regulatory functions of IIAGlc depend on its phosphorylation state, which in turn depends on IICBGlc transport activity. If IICBGlc is not actively transporting glucose, IIAGlc is phosphorylated. If IICBGlc is actively transporting glucose, IIAGlc is mainly dephosphorylated and dephospho-IIAGlc is responsible for inducer exclusion. Dephospho-IIAGlc binds to transporters for other carbon sources, including the lactose permease, and prevents their transport activity. This is one mechanism in wild-type cells that accounts for repression of lactose-inducible genes—e.g., lacZ—when cells are grown in the presence of glucose and lactose. We used this property of the PTS to indirectly measure IICBGlc (PtsG) transport activity. We predicted that if SgrT inhibits glucose transport through PtsG, it should increase levels of phosphorylated IIAGlc and therefore relieve repression of β-galactosidase (LacZ) activity in cells growing on glucose and lactose. A ΔsgrS::kan, lac+ strain carrying vector control or Plac-sgrT plasmids was grown in rich medium with glucose and lactose, and β-galactosidase activity was measured. In cells carrying the vector control, the activity was low (25 Miller units in mid-log phase), reflecting that glucose inhibited uptake of lactose, the inducer and substrate of LacZ (SI Fig. 12). In contrast, cells producing SgrT had dramatically higher levels of β-galactosidase activity (1557 Miller units in mid-log phase) (SI Fig. 12), indicating that SgrT somehow interferes with inducer exclusion. This result further supports our hypothesis that SgrT inhibits glucose transport at the level of PtsG transport activity.
Discussion
Given the relatively large size of SgrS (227 nt), we initially anticipated that SgrS might regulate a large set of mRNA targets similar to other characterized sRNAs; e.g., RyhB, which regulates at least 18 mRNAs by base pairing (19). Instead, the studies presented here suggest an alternative function for SgrS. We identified a conserved ORF in the 5′ region of SgrS that we designated sgrT. Genetic and biochemical data support the notion that the SgrT polypeptide itself has a distinct function that can promote recovery from stress and negatively affect glucose transport. Although the phenotypes for base pairing-only or sgrT-only alleles are the same—i.e., growth rescue (Fig. 3A), growth inhibition on glucose (Fig. 3B), and glucose uptake inhibition (Fig. 6B)—our data indicate that different mechanisms are used to effect these outcomes. The riboregulation function acts through promoting ptsG mRNA degradation (Fig. 5) (4, 6) and inhibiting synthesis of PtsG (Fig. 6A) (7). SgrT does not affect levels of ptsG mRNA or PtsG protein (Figs. 5 and 6A) yet still inhibits glucose uptake (Fig. 6B). Together, these data suggest that the two functions encoded on SgrS have some physiological redundancy.
We propose that the small RNA SgrS encodes two distinct functions that participate in the glucose phosphate stress response. Our previous studies (6), as well as those of Aiba and coworkers (12), have shown that SgrS base pairing activity is required for down-regulation of ptsG mRNA stability and for preventing new synthesis of IICBGlc proteins under stress conditions. Stopping new synthesis of glucose transporters may not be sufficient for relief from stress under conditions where preexisting glucose transporters remain competent for glucose uptake because phosphosugars would continue to accumulate. The identification of SgrT and the evidence for its role in inhibiting glucose transporter activity suggest a mechanism to overcome this problem. It is remarkable that the 227-nt SgrS molecule apparently carries out dual strategies, riboregulation and coding for a protein inhibitor of glucose transport, that reduce uptake of glucose-phosphate under conditions where it is not appropriately metabolized. Inhibition of glucose transport at the level of IICBGlc (PtsG) activity by SgrT might be mediated by protein–protein interactions that result in “plugging” the transport channel or inhibiting IICBGlc phosphorylation. A prediction of this model is that SgrT mediates a rapid response to stress that reduces influx of sugar phosphates without affecting levels or stability of the transport protein. The riboregulation activity of SgrS might normally be an important adaptation to prolonged stress, because it stops synthesis of new transporters, and extant transporters are diluted out as cells continue to grow (Fig. 6A).
In the past few years, sRNA regulators have garnered a great deal of attention and study, yet we are still discovering novel physiological functions and mechanisms of action. By far the most well studied class of sRNAs are those that act by base pairing with target messages, like the microRNAs in eukaryotes. The analogous bacterial sRNAs also act as base pairing-dependent riboregulators that require the action of protein cofactors, most notably Hfq. The majority of characterized bacterial sRNAs base pair with sequences in the 5′ UTRs of their target messages and down-regulate their translation and/or stability. However, new variations on this theme are emerging. Some riboregulators positively regulate translation of their target messages, and there are now examples of riboregulators that act negatively on some targets and positively on others. Our work has now provided evidence for yet another functional class of sRNAs. SgrS and RNA III of S. aureus are bifunctional sRNAs that can act through a base pairing mechanism on mRNA targets and also serve as mRNAs themselves. Although SgrS shares the basic feature of bifunctionality with RNA III, there are some important and intriguing differences. For example, Hfq is required for riboregulation by SgrS but apparently is not required for RNA III. Furthermore, SgrS is thus far unique in that the function of the encoded protein product (SgrT) appears to be redundant physiologically (although not mechanistically) with the riboregulation function. One of the lessons that may be taken from this study is that small ORFs encoded by sRNAs should be examined carefully for potential function. Undoubtedly there are other bifunctional sRNAs that await characterization.
Methods
Strain and Plasmid Construction.
The strains used in this study are listed in SI Table 1, and oligonucleotides are listed in SI Table 2. Alleles were moved between strains by P1 transduction or inserted via λ Red recombination (20). Strains DH5α (Invitrogen) and XL10 gold (Stratagene) were used for cloning procedures. Plasmid vectors were pBR322 derivatives, pHDB3 (21) or pBRPlac (22).
Strains BH300 and BH301 were created by using a modified λ Red and FLP-mediated recombination protocol (23). Briefly, a kanamycin cassette flanked by FRT sites was amplified from template pKD13 (23) by using primers O-CV247 and O-CV248 and integrated into the chromosome by λ Red recombination at the 3′ end of sgrT. The remaining steps were as described in ref. 23. The resulting strains, BH300 (wild-type sgrT) and BH301 (sgrTUAA), carry in-frame translational fusions between sgrT and lacZ. Strain BH302 was created by insertion of the ΔsgrR::cat allele into strain BH300 by λ Red recombination.
Plasmid pLCV1 carries Plac-sgrS and is described in ref. 6. Plasmid pLCV5 was derived from pLCV1 by whole-plasmid PCR mutagenesis using primers O-CV111 and O-CV112. pLCV5 (Plac-sgrSUAA) contains a single base pair substitution that changed the fifth codon of sgrT from “TAT” to “TAA” to create a stop codon. The sgrT ORF was amplified with forward primer O-CV115, containing an AatII restriction site and heterologous 5′ leader and ribosome binding site, and reverse primer O-CV116, which had an EcoRI restriction site. This PCR product was cloned into the vector pBRPlac (22), resulting in plasmid pBRCV7 (Plac-sgrT). Plasmid pBRCV8 (Plac-sgrTUAA) was derived from pBRCV7 and was constructed by PCR mutagenesis using primers O-CV118 and O-CV119 to incorporate the stop codon mutation as described for pLCV5. Plasmids pBRCS1 and pBRCS4 were created by PCR mutagenesis on pBRCV7 and pBRCV8, respectively, using primers O-CV207 and O-CV208. pBRCS1 and pBRCS4 carry additional sequences that encode a 3XFLAG tag fused to the C terminus of SgrT.
Media and Reagents.
LB medium was used for all liquid cultures and plates unless otherwise noted. Media were supplemented with 100 μg/ml ampicillin, 10 μg/ml chloramphenicol, or 25 μg/ml tetracycline where indicated. IPTG was used at a concentration of 0.1 mM for induction of Plac-sgrS or Plac-sgrT alleles and at a concentration of 1 mM for induction of pBRCS1 and pBRCS4 for protein extraction. MOPS EZ Rich defined medium (Teknova) or minimal A medium with 0.2% glucose or 0.4% glycerol was used for some experiments.
Phenotypic Assays.
αMG rescue.
Strains were grown overnight in LB with ampicillin and 0.1 mM IPTG and then subcultured 1:500 in fresh medium. Cultures were grown to early-log phase (OD600 ∼ 0.1), and stress was induced by the addition of 0.5% αMG to the medium. Growth was monitored by measuring the optical density at 600 nm (OD600) every 30 min before addition of αMG and every 20 min thereafter until cells reached stationary phase.
Glucose growth inhibition.
The cultures were grown overnight in minimal A medium with ampicillin, IPTG, and glycerol. Strains were subcultured 1:200 in fresh medium with ampicillin, IPTG, and glucose. Growth was monitored by measuring OD600 every hour until cells reached stationary phase.
Glucose uptake assays.
Cultures were grown overnight in rich defined medium (MOPS EZ Rich) with ampicillin, IPTG, and glycerol. Strains were subcultured 1:500 in fresh medium with ampicillin, IPTG, and glucose. Samples were taken immediately after subculture and again after 5 h of growth. Cells were pelleted by centrifugation, and a 10-fold dilution of supernatant was used with the glucose (HK) assay kit (Sigma), according to the manufacturer's instructions, to determine the amount of glucose in supernatant samples. Protein samples were harvested by precipitation with trichloroacetic acid (TCA) at the 5-h time point. Processing of protein samples and Western blotting is described below.
β-Galactosidase assays.
Strains containing a translational sgrT′-′lacZ fusion were grown overnight in TB medium and subcultured 1:200 to fresh medium. Cultures were grown to OD600 ∼ 0.5 and then split into two cultures, one of which was induced with 0.005% αMG. Samples were taken as indicated after induction and assayed for β-galactosidase activity as described in ref. 24.
Inducer exclusion.
Strains were grown overnight in TB with ampicillin and subcultured 1:200 in fresh media with antibiotic, IPTG, 0.2% glucose, and 0.2% lactose. Samples were taken as indicated, roughly to OD600 0.1, 0.5, and 2, respectively, and were used for Miller assay (24).
RNA Methods.
RNA was extracted by the hot phenol method as described in ref. 25. The concentration of RNA samples was determined spectrophotometrically, and samples were prepared for electrophoresis by using equal amounts of total RNA (15 μg for ptsG Northern blots). Samples were run on 1.2% agarose gels alongside the Millennium size marker (Ambion) at 90 V for ≈2 h. The gel was prepared and RNA transferred as described in ref. 26. Prehybridization was performed in ULTRAhyb (Ambion) solution at 42°C for at least 30 min; the membrane was probed overnight with a 5′-biotinylated probe, RyaA-1bio or ptsG-1bio, for SgrS and ptsG mRNA, respectively. Detection was performed according to BrightStar BioDetect kit (Ambion) specifications.
Protein Methods.
For Western blot analysis, strains were grown overnight in LB with ampicillin and subcultured 1:500 in fresh media. The cultures were grown to mid-log phase (OD600 ∼ 0.5) and induced with IPTG. Proteins were harvested by precipitation with TCA, as described in ref. 17, at the time points indicated; the OD600 was measured when the samples were harvested. All proteins were resuspended in sample buffer with DTT (New England Biolabs).
All protein gels and buffers were from Invitrogen. Protein samples were run on 10% Bis-Tris gels with MOPS-SDS buffer for PtsG or 4–12% Bis-Tris gels with Mes-SDS buffer for SgrT-3XFLAG at the recommended running times and voltages. The proteins were transferred to Immobilon membranes (Millipore): Immobilon-P or Immobilon-PSQ for PtsG (IICBGlc) and SgrT-3XFLAG, respectively. Membranes were blocked and Western blots performed as described in ref. 17. The rabbit primary antibody against PtsG was a gift from Hiroji Aiba (Nagoya University, Nagoya, Japan) and was used at a dilution of 1:5,000, and goat anti-rabbit IgG horseradish peroxidase conjugate secondary antibody (used at 1:5,000) was from Calbiochem. The ECL Plus reagent (GE Healthcare) was used for detection. The autoradiography film used was Kodak BioMax light film.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Hiroji Aiba for providing the anti-IIBGlc antibody and Susan Gottesman, Gisela Storz, Jennifer Han, and Yan Sun for useful discussions and critical reading of the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 20149.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708102104/DC1.
References
- 1.Englesberg E, Anderson RL, Weinberg R, Lee N, Hoffee P, Huttenhauer G, Boyer H. J Bacteriol. 1962;84:137–146. doi: 10.1128/jb.84.1.137-146.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kadner RJ, Murphy GP, Stephens CM. J Gen Microbiol. 1992;138:2007–2014. doi: 10.1099/00221287-138-10-2007. [DOI] [PubMed] [Google Scholar]
- 3.Yarmolinsky MB, Wiesmeyer H, Kalckar HM, Jordon E. Proc Natl Acad Sci USA. 1959;45:1786–1791. doi: 10.1073/pnas.45.12.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kimata K, Tanaka Y, Inada T, Aiba H. EMBO J. 2001;20:3587–3595. doi: 10.1093/emboj/20.13.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morita T, El-Kazzaz W, Tanaka Y, Inada T, Aiba H. J Biol Chem. 2003;278:15608–15614. doi: 10.1074/jbc.M300177200. [DOI] [PubMed] [Google Scholar]
- 6.Vanderpool CK, Gottesman S. Mol Microbiol. 2004;54:1076–1089. doi: 10.1111/j.1365-2958.2004.04348.x. [DOI] [PubMed] [Google Scholar]
- 7.Morita T, Mochizuki Y, Aiba H. Proc Natl Acad Sci USA. 2006;103:4858–4863. doi: 10.1073/pnas.0509638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Mol Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 9.Massé E, Majdalani N, Gottesman S. Curr Opin Microbiol. 2003;6:120–124. doi: 10.1016/s1369-5274(03)00027-4. [DOI] [PubMed] [Google Scholar]
- 10.Møller T, Franch T, Hojrup P, Keene DR, Bachinger HP, Brennan R, Valentin-Hansen P. Mol Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 11.Sledjeski DD, Whitman C, Zhang A. J Bacteriol. 2001;183:1997–2005. doi: 10.1128/JB.183.6.1997-2005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawamoto H, Koide Y, Morita T, Aiba H. Mol Microbiol. 2006;61:1013–1022. doi: 10.1111/j.1365-2958.2006.05288.x. [DOI] [PubMed] [Google Scholar]
- 13.Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, et al. Genes Dev. 2007;21:1353–1366. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huntzinger E, Boisset S, Saveanu C, Benito Y, Geissmann T, Namane A, Lina G, Etienne J, Ehresmann B, Ehresmann C, et al. EMBO J. 2005;24:824–835. doi: 10.1038/sj.emboj.7600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morfeldt E, Taylor D, von Gabain A, Arvidson S. EMBO J. 1995;14:4569–4577. doi: 10.1002/j.1460-2075.1995.tb00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bohn C, Rigoulay C, Bouloc P. BMC Microbiol. 2007;7:10. doi: 10.1186/1471-2180-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vanderpool CK, Gottesman S. J Bacteriol. 2007;189:2238–2248. doi: 10.1128/JB.01689-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deutscher J, Francke C, Postma PW. Microbiol Mol Biol Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Massé E, Vanderpool CK, Gottesman S. J Bacteriol. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu DG, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. Proc Natl Acad Sci USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ulbrandt ND, Newitt JA, Bernstein HD. Cell. 1997;88:187–196. doi: 10.1016/s0092-8674(00)81839-5. [DOI] [PubMed] [Google Scholar]
- 22.Guillier M, Gottesman S. Mol Microbiol. 2006;59:231–247. doi: 10.1111/j.1365-2958.2005.04929.x. [DOI] [PubMed] [Google Scholar]
- 23.Ellermeier CD, Janakiraman A, Slauch JM. Gene. 2002;290:153–161. doi: 10.1016/s0378-1119(02)00551-6. [DOI] [PubMed] [Google Scholar]
- 24.Miller JH. Experiments in Bacterial Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1972. [Google Scholar]
- 25.Aiba H, Adhya S, de Crombrugghe B. J Biol Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 26.Majdalani N, Chen S, Murrow J, St. John K, Gottesman S. Mol Microbiol. 2001;39:1382–1394. doi: 10.1111/j.1365-2958.2001.02329.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.