Abstract
Gap junction channels are required for normal cardiac impulse propagation, and gap junction remodeling is associated with enhanced arrhythmic risk. Oculodentodigital dysplasia (ODDD) is a multisystem syndrome due to mutations in the connexin43 (Cx43) gap junction channel gene. To determine the effects of a human connexin channelopathy on cardiac electrophysiology and arrhythmogenesis, we generated a murine model of ODDD by introducing the disease-causing I130T mutant allele into the mouse genome. Cx43 abundance was markedly reduced in mutant hearts with preferential loss of phosphorylated forms that interfered with trafficking and assembly of gap junctions in the junctional membrane. Dual whole-cell patch–clamp studies showed significantly lower junctional conductance between neonatal cell pairs from mutant hearts, and optical mapping of isolated-perfused hearts with voltage-sensitive dyes demonstrated significant slowing of conduction velocity. Programmed electrical stimulation revealed a markedly increased susceptibility to spontaneous and inducible ventricular tachyarrhythmias. In summary, our data demonstrate that the I130T mutation interferes with Cx43 posttranslational processing, resulting in diminished cell–cell coupling, slowing of impulse propagation, and a proarrhythmic substrate.
Keywords: arrhythmia, connexin43, transgenic, channel, mouse
Gap junction channels are required for normal cardiac impulse propagation, and abnormalities in the expression of such channels, a process known as gap junction remodeling, are associated with enhanced arrhythmic risk (1). Oculodentodigital dysplasia (ODDD) is a multisystem disorder due to mutations in the connexin43 (Cx43) gap junction channel gene, characterized by craniofacial and limb dysmorphism, neurological symptoms, and infrequent cardiac abnormalities, including arrhythmias (2). Using exogenous expression systems, several groups have explored the cellular phenotypes arising from ODDD disease-causing Cx43 mutations. These studies demonstrate a range of abnormalities, including mutant proteins that fail to traffic normally to the junctional membrane and others that traffic efficiently but nonetheless have decreased channel function. In some cases, the mutant protein dominantly inhibits channel activity when coexpressed with wild-type Cx43 (3–8), consistent with the autosomal dominant mode of inheritance of ODDD and reminiscent of the behavior of a subset of potassium channel mutants responsible for long QT syndrome (9). To date, none of the naturally occurring ODDD mutations have been examined in vivo, although a recent report indicated that a glycine-to-serine missense mutation at residue 60 within the highly conserved first extracellular loop of Cx43 recapitulated some features of ODDD (10). To characterize the molecular pathogenesis of an inherited Cx43 gap junction channelopathy, we developed an in vivo model of ODDD by introducing the human disease-causing I130T mutation into the murine genome. Family members harboring this mutation, which resides in the cytoplasmic loop domain, appear to have an increased incidence of cardiac rhythm disturbances (2). Accordingly, our analysis focused on electrophysiological manifestations in this model.
Results
Survival Disadvantage in ODDD Mutant Mice.
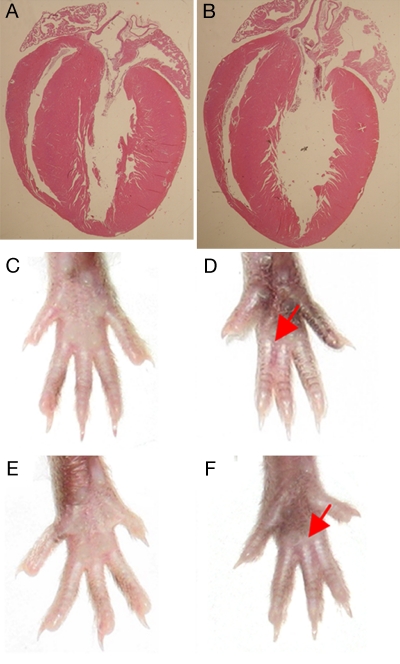
To explore the molecular pathogenesis of ODDD and to determine the effects of the disease-causing I130T Cx43 mutation on cardiac impulse propagation and arrhythmogenesis, we established a murine model of this syndrome, using the targeting strategy shown in Fig. 1. Northern blot analysis demonstrated equivalent levels of Cx43 mRNA abundance in the hearts of heterozygous mutant mice and wild-type littermate controls, indicating that the targeting event did not interfere with expression at the messenger RNA level (Fig. 1E). Mutant mice displayed a survival disadvantage, because significantly fewer than the Mendelian proportion of neonatal heterozygous Cx43I130T/+ mice (36%, 76 of 210) were produced from matings between heterozygotes and wild types (χ2 = 16.0; P < 0.00001), and no Cx43I130T/I130T homozygotes (0%, 0 of 77) were identified in crosses between heterozygous mating pairs. Histological examination of adult Cx43I130T/+ hearts (n = 5) did not reveal any of the morphological abnormalities reported in Cx43+/− germ-line knockout (11, 12) or Cx43Jrt/+ G60S mutant mice (10), such as right ventricular outflow obstruction, atrial septal defect, or patent foramen ovale, and echocardiographic assessment of LV structure and contractile function showed no significant differences between adult Cx43I130T/+ mutant mice and littermate controls (Fig. 2 A and B). The basis for the embryonic mortality in mutant mice was not pursued further in this study. Interestingly, almost all (93%, 71 of 76) Cx43I130T/+ mice displayed hind-limb syndactyly, a characteristic phenotypic feature observed in human ODDD patients (Fig. 2 C–F).
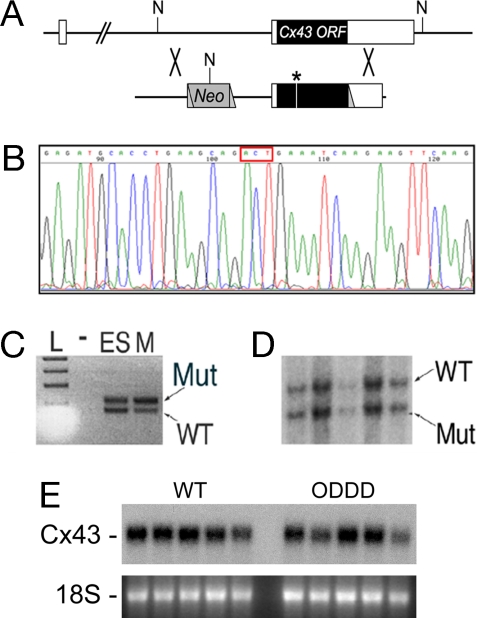
Fig. 1.
Generation of Cx43I130T/+ Mutant mice. (A) The Cx43 locus (Upper) and targeting vector (Lower) are shown. The locus comprises two exons with the ORF within exon 2. The neomycin (Neo) resistance cassette, loxP sites (triangles), and location of the I130T mutation (asterisk) within the targeting vector are shown. NcoI restriction sites (N) are indicated. (B) Sequence analysis of targeting vector demonstrating introduction of the I130T missense mutation (ACT codon in red box). (C) PCR assay with primer pairs that flank the loxP site residing in the 3′ untranslated region of the targeted allele. Lanes include ladder (L), no input DNA (−), DNA from heterozygous targeted ES cells (ES), or DNA from an F1 mutant mouse (M). PCR products indicative of both the wild-type (WT) and mutant (Mut) I130T alleles are present. (D) Southern blot analysis of NcoI-digested genomic DNA from F2 offspring positive for heterozygosity by PCR assay, showing the expected polymorphism and confirming transmission of both the wild-type and mutant alleles. (E) Northern blot analysis of wild-type (WT) and ODDD mutant hearts demonstrate comparable expression of Cx43. Ethidium bromide staining of 18S ribosomal RNA is also shown.
Fig. 2.
Cardiac and limb morphology in ODDD mutant mice. (A and B) Four-chamber view of 3-month-old wild-type (A) and ODDD mutant (B) hearts demonstrates preserved cardiac morphology. (C–E) Comparison of left (C and D) and right (E and F) hind limbs from wild-type and heterozygous ODDD mutant mice. Syndactyly in mutants is indicated by red arrows.
Aberrant Phosphorylation and Trafficking of Cx43 in ODDD Mutant Hearts.
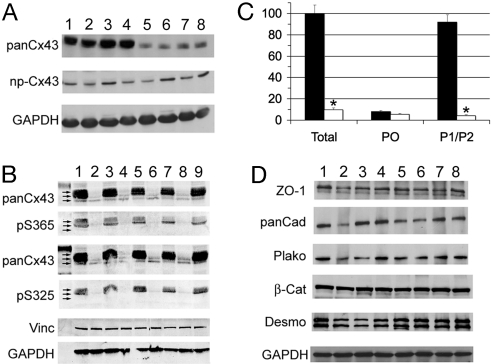
Western blot analysis demonstrated that the abundance of total Cx43 protein in hearts from heterozygous ODDD mutant mice was significantly reduced, to 9.7 ± 2.3% of the levels in wild-type littermate controls (wild type, 100 ± 9.0; ODDD, 9.7 ± 2.3; P < 0.0001), consistent with a dominant-negative effect of the mutant protein on the stability of wild-type Cx43 (Fig. 3A). Interestingly, the abundance of the P0 variant of Cx43 was not significantly different in wild-type and ODDD mutant hearts (Fig. 3A), and the difference in total Cx43 was accounted for entirely by a specific reduction in the processed phosphorylated P1 and P2 forms in ODDD mutant hearts (Fig. 3B), as determined with antibodies specific for phospho365-Cx43 (P1) and phospho325/328/330-Cx43 (P2). These data are summarized in Fig. 3C.
Fig. 3.
Aberrant expression of Cx43 in ODDD mutant hearts. (A) Western blot analysis of wild-type (WT) and ODDD mutant (ODDD) hearts using rabbit antibody 18B, which reacts with all forms of Cx43 (panCx43) and mouse monoclonal CX1B1, which preferentially reacts with the P0 form of Cx43 (np-Cx43). Lanes 1–4 are from wild-type hearts, and lanes 5–8 are from ODDD hearts. Signals were visualized with enhanced chemiluminescence. (B) Western blot analysis using mouse monoclonal Cx43NT1, which recognizes all forms of Cx43, and rabbit anti-pS325/328/330-Cx43 and rabbit anti-pS365-Cx43 antibodies, which recognize specific phosphorylated forms of Cx43. Lanes 1, 3, 5, 7, and 9 are from wild-type hearts, and lanes 2, 4, 6, and 8 are from ODDD hearts. Equivalency of loading was verified by probing for vinculin (vinc) and GAPDH. Signals were visualized and quantified using the Li-Cor imaging system. Arrows in A and B indicate P0, P1, and P2 forms of Cx43. (C) Abundance of total Cx43, P0, and P1/P2 forms of Cx43 in wild-type (■) and ODDD (□) hearts. Values are expressed as mean ± SEM relative to total Cx43 levels in wild-type hearts. (D) Western blot analysis for zona occludens-1 (ZO-1), pan-cadherin (panCad), plakoglobin (plako), β-catenin (β-Cat), desmoplakin (desmo), and GAPDH. Lanes 1–4 are from wild-type hearts, and lanes 5–8 are from ODDD hearts.
Expression of connexin45 and connexin40 was not statistically different in mutant and wild-type hearts (data not shown), nor were a panel of other junctional proteins with the exception of desmoplakin, which showed a 36% increase in the mutant hearts compared with controls (wild type, 100 ± 10.0% (n = 4); ODDD, 136 ± 6.0% (n = 4); P = 0.014), as shown in Fig. 3D.
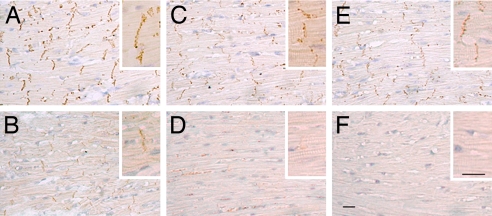
The consequences of aberrant biochemical processing of Cx43 on gap junctional formation were evaluated by immunohistochemical staining. Compared with wild-type controls (Fig. 4A), there was a marked reduction in accumulation of Cx43 at the junctional membrane, as visualized with antibodies recognizing all forms of Cx43 (Fig. 4B). Moreover, consistent with Western blot analyses showing reductions in posttranslationally modified P1 and P2 isoforms, immunohistochemical staining with these same phospho-specific Cx43 antibodies demonstrated a dramatic loss of phosphorylation at serines S325/328/330 (Fig. 4 C and D) as well as S365 (Fig. 4 E and F). Phosphorylation of Cx43 at S365 is involved in gap junction assembly and formation of the P1 isoforms, whereas phosphorylation at S325/328/330 is involved in P2 formation and thought to be a target of casein kinase 1δ (13, 14). The parallel acute loss of total Cx43, pS325/328/330, and pS365 indicates reduced formation of gap junctional structures at intercalated disks. We also examined phosphorylation at the PKC-specific site S368; the level of staining was low in wild type and absent in ODDD hearts (data not shown).
Fig. 4.
Cx43 gap junctions are diminished in ODDD mutant hearts. Immunohistochemical staining of wild-type (A, C, and E) and ODDD mutant hearts (B, D, and F) with antibodies recognizing all forms of Cx43 (A and B), phosphoS325/328/330-Cx43 (C and D), and phosphoS365-Cx43 (E and F). Phosphorylated forms of Cx43 are virtually absent in ODDD mutant hearts. (Scale bar: 20 μm.)
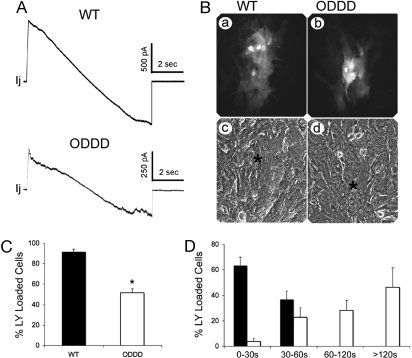
Junctional Conductance and Lucifer Yellow (LY) Dye Transfer Is Reduced in Cells from ODDD Mutant Hearts.
Dephosphorylation of Cx43 in cardiomyocytes has been associated with cellular uncoupling. Accordingly, we next evaluated junctional conductance in cell pairs from neonatal hearts of ODDD mutant mice and littermate controls. Consistent with this notion, junctional conductance was significantly reduced in ODDD mutant cell pairs compared with wild-type controls (wild type, 15.7 ± 1.4 nS (n = 19); ODDD, 7.9 ± 2.4 nS (n = 11); P = 0.006), as summarized in Table 1. Junctional currents recorded in response to transjunctional voltage ramps from representative wild-type and ODDD heterozygote cardiac myocyte pairs are shown in Fig. 5A; voltage dependence of junctional currents in both genotypes is apparent in junctional currents at the highest transjunctional voltages. Interestingly, the extent of reduction in coupling between heterozygous ODDD mutant cell pairs is similar to that reported for Cx43−/− myocytes from germ-line knockout mice (15). In addition, intercellular coupling was evaluated by LY dye injections. Consistent with the in vitro studies of Lai et al. (8) showing reduced sulforhodamine B dye transfer in transfected cells coexpressing wild-type Cx43 and eYFP-tagged I130T mutant Cx43, we found that LY dye transfer in cardiac myocytes from ODDD heterozygotes (which coexpress wild-type and mutant Cx43) was significantly reduced compared with wild-type controls [percent LY loaded cells; wild type, 91.6 ± 2.6 (n = 45 cells); ODDD, 51.6 ± 4.2 (n = 85 cells); P < 0.0001]. On average, dye transfer was six times slower in ODDD mutants compared with wild-type controls [wild type, 25.1 ± 1.9 sec; (n = 30 cells), ODDD, 150.5 ± 13.3 sec (n = 61 cells); P < 0.0001]. These data are summarized in Fig. 5 B–D.
Table 1.
Electrophysiological characterization of wild-type and ODDD mutant mice
| Wild type | ODDD | P | |
|---|---|---|---|
| Junctional conductance | |||
| Gj, nS | 15.7 ± 1.4 (19) | 7.9 ± 2.4 (11) | 0.006 |
| Surface ECG | |||
| RR, ms | 139 ± 3.8 (8) | 134 ± 5.0 (7) | 0.46 |
| PR, ms | 40.3 ± 1.8 (8) | 42.3 ± 1.8 (7) | 0.46 |
| QRS, ms | 10.5 ± 0.3 (8) | 12.3 ± 1.0 (7) | 0.10 |
| QT, ms | 49.8 ± 1.9 (8) | 55.6 ± 5.6 (7) | 0.32 |
| QTc | 42.2 ± 1.4 (8) | 47.9 ± 4.3 (7) | 0.21 |
| R/P ratio | 9.05 ± 0.88 (3) | 2.83 ± 0.63 (4) | <0.002 |
| Optical mapping | |||
| CVmax, m/s | 0.85 ± 0.03 (6) | 0.64 ± 0.04 (8) | <0.005 |
| CVmin, m/s | 0.48 ± 0.03 (6) | 0.34 ± 0.02 (8) | <0.005 |
Gj, junctional conductance. Number in parentheses indicates sample size.
Fig. 5.
Coupling is reduced in ODDD mutant myocytes. (A) Junctional currents recorded between pairs of cardiac myocytes isolated from wild-type (Upper) and ODDD heterozygotes (Lower). Both cells were voltage-clamped with patch-type electrodes and held at 0 mV; junctional current (Ij) was recorded in one cell when the other's voltage was ramped from −100 to 100 mV at 27 mV/sec. Note nonlinearities in Ij at large transjunctional voltages for both genotypes, due to voltage-dependent gating of junctional channels. (B) Representative examples of Lucifer yellow dye coupling in cultures of wild-type (a and c) and heterozygous ODDD mutant (b and d) myocytes. (C) Quantification of dye transfer assay shows significantly reduced coupling in ODDD cultures compared with wild-type (WT) cultures. (D) The rate of dye transfer was significantly slower between ODDD mutant myocytes (ρ) compared with wild type (ν).
Impulse Propagation Is Slowed and Arrhythmia Propensity Is Increased in ODDD Mutants.
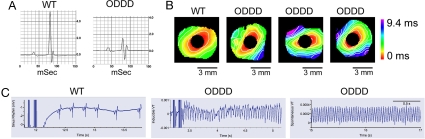
We next determined whether the reduction in cell–cell coupling was associated with alterations in cardiac impulse propagation. Baseline surface ECGs in lightly anesthetized mice were performed as described (16, 17), and results are summarized in Table 1. We observed no differences in heart rate, PR interval, or QT interval corrected for heart rate (QTc). There was a trend toward prolongation of the QRS interval, but this did not reach statistical significance, although there are limitations associated with surface electrocardiography, as discussed (18). Interestingly, mutant mice displayed diminished amplitude of the QRS complex (Fig. 6A), a phenomenon similar to that observed in Cx43 conditional knockout mice (16). The ratio of R wave/P wave amplitude was significantly reduced in ODDD mutant mice, as shown in Table 1.
Fig. 6.
Abnormal conduction and arrhythmias in ODDD mutant mice. (A) Representative signal-averaged surface electrocardiograms (lead II) from a wild-type (WT) and an ODDD mutant mouse. Note the diminished QRS amplitude in the mutant. (B) Optical mapping of the left ventricular surface of a representative wild-type heart and three individual ODDD mutant hearts. The temporal and spatial scales are indicated and are identical for all images. Conduction velocity, determined from pixels between 1 and 3 mm from the site of stimulation, is significantly slowed in the ODDD hearts, as indicated by closer spacing of isochrones (1 ms apart). (C) Programmed electrical stimulation showing examples of return of sinus rhythm in a wild-type heart (Left), induction of sustained VT in an ODDD heart (Center), and spontaneous VT in an ODDD heart (Right). Tracings each include 2 sec of recordings.
To assess conduction properties more directly, left ventricular epicardial conduction velocities were determined by optical recordings in isolated-perfused hearts using voltage-sensitive dyes. We observed that conduction velocity in both the longitudinal (CVmax) and transverse directions (CVmin) was significantly slowed compared with wild-type controls (Fig. 6B and Table 1). Inasmuch as conduction slowing is a major contributor enhancing the propensity for reentrant ventricular arrhythmias, we also performed provocative programmed electrical stimulation to assess arrhythmic potential in mutant hearts and littermate controls. All eight ODDD mutant hearts displayed inducible (six of eight) or spontaneous (two of eight) sustained ventricular tachycardia (VT), whereas no wild-type hearts had sustained arrhythmias (χ2 = 14; P < 0.001) (Fig. 6C).
In summary, we have developed a murine model of an inherited connexin channelopathy. Our data indicate that aberrant posttranslational processing of the mutant Cx43 is associated with diminished trafficking and assembly of junctional channels, reduced intercellular coupling, slow conduction, and enhanced arrhythmic potential. The markedly reduced steady-state abundance of Cx43 indicates that the mutant protein exerts a dominant-negative effect on wild-type Cx43, presumably through oligomerization of mutant and wild-type monomers and degradation of the resulting complex. This behavior is similar to that observed in Cx43Jrt/+ mutant mice (10) and is consistent with the autosomal-dominant mode of inheritance of ODDD. In addition to changes in gap junction abundance at the intercalated discs, the aberrant posttranslational phosphorylation of Cx43 may influence the gating properties of assembled gap junction channels, as has been reported by several groups (19–21) and reviewed in refs. 20 and 22. Although the relative contribution of each of these mechanisms to the diminished cell–cell coupling we observe will require further study, our functional analysis of junctional conductance indicate that cell–cell coupling in ODDD cell pairs approximates that observed in Cx43-null myocytes from germ-line knockout mice (15).
Interestingly, we did not observe overt structural defects in the hearts of Cx43I130T/+ mice, unlike the Cx43Jrt/+ mutant mice reported by Rossant and colleagues (11). The G60S mutation identified in those mice resulted from an unbiased chemical mutagenic screen, which may facilitate the recovery of phenotypes more severe than naturally occurring human mutations such as the I130T missense mutation reported in this study.
Importantly, the molecular pathology in Cx43I130T/+ mutant mice appears similar to derangements observed in acquired gap junction remodeling, in which hypophosphorylation and diminished accumulation of functional channels at the intercalated disk are stereotypical findings (1). These mechanistic similarities suggest that the I130T ODDD mutant mice may serve as a useful platform to test therapies to ameliorate the functional consequences of gap junction remodeling.
Methods
Mice.
Site-directed mutagenesis was performed to introduce the disease-causing I130T point mutation into the same gene-targeting vector used to conditionally inactivate Cx43 in the heart and other lineages (23–25). Targeting was performed in the 129/Sv-derived ES cell line R1 (23, 26). Screening for correct recombinants was performed by Southern blot and PCR analysis, also as described (10). One correctly targeted ES clone was injected into C57BL/6 blastocysts. Highly chimeric male mice were crossed with wild-type CD1 females to generate F1 Cx43I130T/+ heterozygous mutant mice. For all experiments, wild-type littermates were used as controls. All experiments were performed in accordance with the regulations of our Institutional Animal Care and Use Committees.
Western Blot Analysis.
Cardiac lysates were prepared from 20- to 25-week-old animals, and Western blots were performed as described with a panel of antibodies to connexins, phosphospecific forms of Cx43, and a panel of junction-associated proteins, as detailed in Fig. 3 and refs. 13, 27, and 28. The level of antibody binding was quantified by using the Li-Cor imaging system, and loading was normalized to GAPDH.
Immunohistological Analyses.
Staining was performed on formalin-fixed paraffin sections of hearts from wild-type and Cx43I130T/+ mice as described (27, 29).
Dual Whole-Cell Patch–Clamp Analyses and Lucifer Yellow Dye Transfer.
Hearts obtained from individual pups from matings between wild-type and Cx43I130T/+ mice were dissociated and Lucifer yellow dye transfer assays, and dual whole-cell patch–clamp studies were performed as described (15, 30). After genotype was determined, results from wild type and heterozygotes were pooled for statistical analysis.
Electrocardiography.
Surface ECGs were recorded from mice lightly anesthetized with inhaled isoflurane. ECG acquisition was performed by using AcqKnowledge 3.9.1 (Biopac Systems). Thirty seconds of continuous data were signal-averaged, and intervals were determined according to reported methods (16, 17).
Optical Mapping and Arrhythmia Induction.
Optical mapping of voltage-dependent signals in isolated-perfused hearts was performed, and ventricular conduction velocities in the longitudinal (CVmax) and transverse (CVmin) directions were calculated. Pixels near the stimulation site (<1 mm) were excluded to remove any stimulus artifacts and at a distance (>3 mm) to exclude potential wavefront collisions and 3D-wave propagation, as described (23, 31–33). Programmed electrical stimulation to assess arrhythmia inducibility was performed by decremental pacing, starting with a paced cycle length (PCL) of 100 ms for 700 beats and reducing the PCL by 10 ms until either an arrhythmia was induced or capture was lost. The protocol was repeated up to three times if no arrhythmia was induced during the first two runs. Arrhythmias lasting longer than 2 min were considered sustained.
Statistical Analysis.
Data are expressed as mean ± SEM. The t test was used to compare experimental groups, with the exception of survival analysis and arrhythmia inducibility, in which case the c2 test was used. A P value <0.05 was considered statistically significant in all cases.
ACKNOWLEDGMENTS.
We thank Gregory Morley for help with the optical mapping experiments and Lucrecia Marquez-Rosado for help with immunohistochemical studies. This study is supported by National Institutes of Health Grants HL64757 and HL82727 (to G.I.F.), GM55632 (to P.D.L.), and HD32573 (to D.C.S.) and by a Glorney–Raisbeck Fellowship in Cardiovascular Diseases from the New York Academy of Medicine (to N.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Saffitz JE, Schuessler RB, Yamada KA. Cardiovasc Res. 1999;42:309–317. doi: 10.1016/s0008-6363(99)00023-1. [DOI] [PubMed] [Google Scholar]
- 2.Paznekas WA, Boyadjiev SA, Shapiro RE, Daniels O, Wollnik B, Keegan CE, Innis JW, Dinulos MB, Christian C, Hannibal MC, et al. Am J Hum Genet. 2003;72:408–418. doi: 10.1086/346090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seki A, Coombs W, Taffet SM, Delmar M. Heart Rhythm. 2004;1:227–233. doi: 10.1016/j.hrthm.2004.03.066. [DOI] [PubMed] [Google Scholar]
- 4.Shibayama J, Paznekas W, Seki A, Taffet S, Jabs EW, Delmar M, Musa H. Circ Res. 2005;96:e83–e91. doi: 10.1161/01.RES.0000168369.79972.d2. [DOI] [PubMed] [Google Scholar]
- 5.Roscoe W, Veitch GI, Gong XQ, Pellegrino E, Bai D, McLachlan E, Shao Q, Kidder GM, Laird DW. J Biol Chem. 2005;280:11458–11466. doi: 10.1074/jbc.M409564200. [DOI] [PubMed] [Google Scholar]
- 6.McLachlan E, Manias JL, Gong XQ, Lounsbury CS, Shao Q, Bernier SM, Bai D, Laird DW. Cell Commun Adhes. 2005;12:279–292. doi: 10.1080/15419060500514143. [DOI] [PubMed] [Google Scholar]
- 7.Gong XQ, Shao Q, Lounsbury CS, Bai D, Laird DW. J Biol Chem. 2006;281:31801–31811. doi: 10.1074/jbc.M605961200. [DOI] [PubMed] [Google Scholar]
- 8.Lai A, Le DN, Paznekas WA, Gifford WD, Jabs EW, Charles AC. J Cell Sci. 2006;119:532–541. doi: 10.1242/jcs.02770. [DOI] [PubMed] [Google Scholar]
- 9.Keating MT, Sanguinetti MC. Cell. 2001;104:569–580. doi: 10.1016/s0092-8674(01)00243-4. [DOI] [PubMed] [Google Scholar]
- 10.Flenniken AM, Osborne LR, Anderson N, Ciliberti N, Fleming C, Gittens JE, Gong XQ, Kelsey LB, Lounsbury C, Moreno L, et al. Development. 2005;132:4375–4386. doi: 10.1242/dev.02011. [DOI] [PubMed] [Google Scholar]
- 11.Reaume AG, de Sousa PA, Kulkarni S, Langille BL, Zhu D, Davies TC, Juneja SC, Kidder GM, Rossant J. Science. 1995;267:1831–1834. doi: 10.1126/science.7892609. [DOI] [PubMed] [Google Scholar]
- 12.Ya J, Erdtsieck-Ernste EB, de Boer PA, van Kempen MJ, Jongsma H, Gros D, Moorman AF, Lamers WH. Circ Res. 1998;82:360–366. doi: 10.1161/01.res.82.3.360. [DOI] [PubMed] [Google Scholar]
- 13.Solan JL, Marquez-Rosado L, Sorgen PL, Thornton PJ, Gafken PR, Lampe PD. J Cell Biol. 2007 doi: 10.1083/jcb.200707060. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper CD, Lampe PD. J Biol Chem. 2002;277:44962–44968. doi: 10.1074/jbc.M209427200. [DOI] [PubMed] [Google Scholar]
- 15.Vink MJ, Suadicani SO, Vieira DM, Urban-Maldonado M, Gao Y, Fishman GI, Spray DC. Cardiovasc Res. 2004;62:397–406. doi: 10.1016/j.cardiores.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell GF, Jeron A, Koren G. Am J Physiol. 1998;274:H747–H751. doi: 10.1152/ajpheart.1998.274.3.H747. [DOI] [PubMed] [Google Scholar]
- 17.Remme CA, Verkerk AO, Nuyens D., van Ginneken AC, van Brunschot S, Belterman CN, Wilders R, van Roon MA, Tan HL, Wilde AA, et al. Circulation. 2006;114:2584–2594. doi: 10.1161/CIRCULATIONAHA.106.653949. [DOI] [PubMed] [Google Scholar]
- 18.Danik S, Cabo C, Chiello C, Kang S, Wit AL, Coromilas J. Am J Physiol. 2002;283:H372–H381. doi: 10.1152/ajpheart.01091.2001. [DOI] [PubMed] [Google Scholar]
- 19.Moreno AP, Saez JC, Fishman GI, Spray DC. Circ Res. 1994;74:1050–1057. doi: 10.1161/01.res.74.6.1050. [DOI] [PubMed] [Google Scholar]
- 20.Kwak BR, van Veen TA, Analbers LJ, Jongsma HJ. Exp Cell Res. 1995;220:456–463. doi: 10.1006/excr.1995.1337. [DOI] [PubMed] [Google Scholar]
- 21.Ek-Vitorin JF, King TJ, Heyman NS, Lampe PD, Burt JM. Circ Res. 2006;98:1498–1505. doi: 10.1161/01.RES.0000227572.45891.2c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lampe PD, Lau AF. Int J Biochem Cell Biol. 2004;36:1171–1186. doi: 10.1016/S1357-2725(03)00264-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gutstein DE, Morley GE, Tamaddon H, Vaidya D, Schneider MD, Chen J, Chien KR, Stuhlmann H, Fishman GI. Circ Res. 2001;88:333–339. doi: 10.1161/01.res.88.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Presley CA, Lee AW, Kastl B, Igbinosa I, Yamada Y, Fishman GI, Gutstein DE, Cancelas JA. Cell Commun Adhes. 2005;12:307–317. doi: 10.1080/15419060500514200. [DOI] [PubMed] [Google Scholar]
- 25.Sridharan S, Simon L, Meling DD, Cyr DG, Gutstein DE, Fishman GI, Guillou F, Cooke PS. Biol Reprod. 2007;76:804–812. doi: 10.1095/biolreprod.106.059212. [DOI] [PubMed] [Google Scholar]
- 26.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gutstein DE, Liu FY, Meyers MB, Choo A, Fishman GI. J Cell Sci. 2003;116:875–885. doi: 10.1242/jcs.00258. [DOI] [PubMed] [Google Scholar]
- 28.Lampe PD, Cooper CD, King TJ, Burt JM. J Cell Sci. 2006;119:3435–3442. doi: 10.1242/jcs.03089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.King TJ, Lampe PD. Cancer Res. 2004;64:7191–7196. doi: 10.1158/0008-5472.CAN-04-0624. [DOI] [PubMed] [Google Scholar]
- 30.del Corsso C, Srinivas M, Urban-Maldonado M, Moreno AP, Fort AG, Fishman GI, Spray DC. Nature Protocols. 2006;1:1799–1809. doi: 10.1038/nprot.2006.266. [DOI] [PubMed] [Google Scholar]
- 31.Danik SB, Liu F, Zhang J, Suk HJ, Morley GE, Fishman GI, Gutstein DE. Circ Res. 2004;95:1035–1041. doi: 10.1161/01.RES.0000148664.33695.2a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morley GE, Danik SB, Bernstein S, Sun Y, Rosner G, Gutstein DE, Fishman GI. Proc Natl Acad Sci USA. 2005;102:4126–4129. doi: 10.1073/pnas.0500881102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamaddon HS, Vaidya D, Simon AM, Paul DL, Jalife J, Morley GE. Circ Res. 2000;87:929–936. doi: 10.1161/01.res.87.10.929. [DOI] [PubMed] [Google Scholar]