Abstract
Protein kinases play a pivotal role in cell signaling, and dysregulation of many kinases has been linked to disease development. A large number of kinase inhibitors are therefore currently under investigation in clinical trials, and so far seven inhibitors have been approved as anti-cancer drugs. In addition, kinase inhibitors are widely used as specific probes to study cell signaling, but systematic studies describing selectivity of these reagents across a panel of diverse kinases are largely lacking. Here we evaluated the specificity of 156 validated kinase inhibitors, including inhibitors used in clinical trials, against 60 human Ser/Thr kinases using a thermal stability shift assay. Our analysis revealed many unexpected cross-reactivities for inhibitors thought to be specific for certain targets. We also found that certain combinations of active-site residues in the ATP-binding site correlated with the detected ligand promiscuity and that some kinases are highly sensitive to inhibition using diverse chemotypes, suggesting them as preferred intervention points. Our results uncovered also inhibitor cross-reactivities that may lead to alternate clinical applications. For example, LY333′531, a PKCβ inhibitor currently in phase III clinical trials, efficiently inhibited PIM1 kinase in our screen, a suggested target for treatment of leukemia. We determined the binding mode of this inhibitor by x-ray crystallography and in addition showed that LY333′531 induced cell death and significantly suppressed growth of leukemic cells from acute myeloid leukemia patients.
Keywords: acute myeloid leukemia, inhibitor selectivity, LY333′531, PIM1, screening data
Reversible protein phosphorylation of tyrosine, threonine, and serine residues is a major control mechanism in mammalian cell signaling. This process is governed by protein kinases, a family of enzymes that comprises 518 members in humans (1). Deregulation of protein kinase activity is associated with a variety of diseases such as cancer and inflammation (2), and the central role of protein kinases in disease pathology has attracted significant interest. In recent years an increasing number of protein kinase inhibitors have entered clinical trials and clinical practice (3, 4). Among the most notable examples is imatinib mesylate (Gleevec), an inhibitor of c-Abl tyrosine kinase (5) that targets an inactive conformation of c-Abl and a few related tyrosine kinases. However, most inhibitors target the active kinase conformation in an ATP mimetic binding mode; hence, because of high active-site similarities of closely related kinases, specificity issues are critical considerations for current inhibitor development to avoid unwanted side-effects caused by inhibition of anti-targets.
To date, substantial inhibition data have been reported in the literature, and even more such data are kept confidential in the private sector. Based on available structural and interaction data, significant efforts have been made to derive information regarding identification of residues required for inhibitor binding and specificity (6–10). However, most published studies have been applied only to a single target or to a small selection of kinases (6). This effort has generated numerous nonoverlapping sets of ligand binding data with largely incomparable assay values. One notable exception is the study by Fabian et al. (11) in which 20 inhibitors derived from a number of unique scaffolds (including 16 inhibitors in clinical trials and approved marketed drugs) have been screened against 119 kinases expressed as viral fusion proteins. In addition, a profiling of 65 inhibitors against 60 kinases has recently been reported (12).
The study presented here provides publicly available data for a set of 156 widely used kinase inhibitors versus a sizeable panel (60 targets) of representative human protein kinases screened by using a systematic and standardized assay. From the data obtained we identified one cross-reacting inhibitor already in phase III clinical trials for which we propose a new application.
Results
The development of kinase inhibitors is one of the key areas for research in pharmaceutical companies as well as in academic science, and a large number of kinase inhibitors are currently used as specific pathway modifiers in cell biology. The study presented here focused on the profiling of a panel of 60 representative human Ser/Thr kinases screened against a diverse set of 156 commercially available kinase inhibitors. This compound set contains at least 16 inhibitors that are or have been in clinical trials or are approved for clinical applications [supporting information (SI) Table 1]. Eleven compounds used in our study were also included in the study by Fabian et al. (11). The location of the profiled kinases in the kinase phylogenetic tree is illustrated in Fig. 1, and an inhibition heat map of our array is shown in Fig. 2. From the selected set of kinases, we have so far been able to determine the structures for catalytic domains from 23 unique proteins at an average resolution of 2.0 Å (SI Table 2). Importantly, 16 of 23 structures were determined in complex with an inhibitor identified from in-house screening, representing valuable starting points for rational ligand design. In addition, we have generated sets of structures for four of these kinases (PIM1, nine structures; SLK, four structures; PAK4, three structures; CLK1, two structures; CK1γ, five structures), with different ligands bound in the active site. Detailed descriptions of each structure and methods used are available at www.sgc.ox.ac.uk/structures/KIN.html.
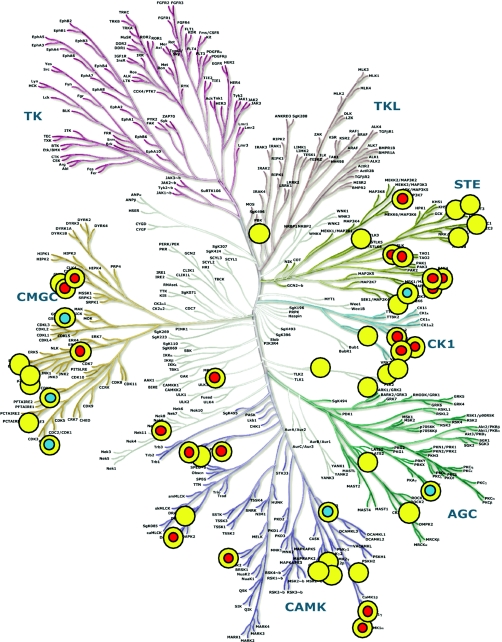
Fig. 1.
Phylogenetic tree of the human protein kinase family (1). Screened targets are indicated by a yellow dot, and structure-determined catalytic domains by the Structural Genomics Consortium or other laboratories are indicated by a red dot and a blue dot with a yellow sphere, respectively. The kinase dendrogram is adapted from ref. 1 and has been reproduced with permission from Science and Cell Signaling.
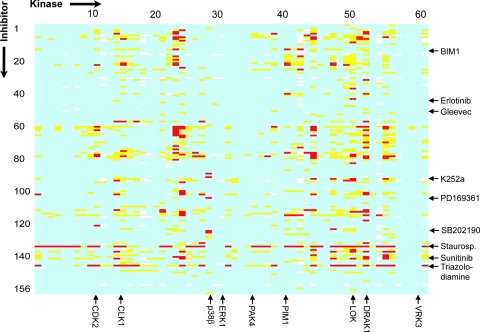
Fig. 2.
Inhibitor array of the screened kinases. Yellow fields indicate a Tm shift >4°C and red field of >8°C, respectively. White fields represent data that have not been determined. A complete table with Tm values is available in SI Table 3. Some targets and inhibitors discussed in the text have been annotated in the figure.
The screening data were generated by using a thermal shift assay in which the kinase is stabilized upon inhibitor binding (13, 14). This simple and cost-effective screening method allowed us to target both active and inactive kinases and has been shown to provide data that are consistent with orthogonal methods such as isothermal titration calorimetry and enzymatic assays (13, 15–17). A comparison of Tm shift data with IC50 values of two targets has been included in SI Fig. 5. The linear correlation of the −log (IC50) versus ΔTm plot suggested that a 4°C shift in thermal stability at an inhibitor concentration 10 μM corresponds to a binding affinity <1 μM. The second threshold in our inhibitor “ranking” was set to 8°C, which typically reflects a Kd of 100 nM or less. In the array shown in Fig. 2 we highlighted only some targets and inhibitors discussed in the article. However, all screening data are compiled in SI Table 3, and compound names, published targets, and suppliers are listed in SI Table 2. From the 156 kinase inhibitors tested, 116 produced Tm shifts above 4°C for at least one kinase in our panel and were thus classified as a significant “hit.”
Inhibitor Selectivity.
A number of inhibitors showed remarkable selectivity in our panel. As expected, the type II inhibitor Gleevec (imatinib) interacted only weakly with PCTK1, adding further validation to its favorable selectivity profile. In the study by Fabian et al. (11) the Gleevec “interactome” was restricted to a small set of kinases, most of them receptor tyrosine kinases together with the Ser/Thr kinases CLK1 and DRAK1, which were also included in our panel. For CLK1 we were not able to identify any significant interaction; however, for DRAK1 the induced stabilization was just below our selected 4°C threshold (Tm = 3.2°C). From approved drugs the tyrosine kinase inhibitors dasatenib and gefitinib interacted only weakly with the Ser/Thr kinases selected in our panel, but the EGFR inhibitor erlotinib (Tarceva) strongly interacted with LOK and the closely related kinase SLK.
Several other inhibitors were confirmed as selective. For example, the specific PKA inhibitor H-89 showed binding only to PKA in our assay. Another example is a group of specific inhibitors of p38α/β, which were developed based on a common scaffold (e.g., PD169316 and SB202190). All of them displayed very good specificity toward p38β as intended (Fig. 2). These and other compounds serve as internal positive controls for the assay method and confirm the validity of data on cross-reactivity for less specific inhibitors identified in this study. In addition, we identified naphthoquinone 1, a selective inhibitor of MEK (IC50 = 0.38 μM for MEK1) (18) and a strong inhibitor of p38β. A number of kinase inhibitors that are non-ATP-competitive (e.g., the CAMKK inhibitor KN-93) or act on kinases indirectly (e.g., geldamycin) were not identified as kinase inhibitors in our screen.
The nonselective inhibitor staurosporine was the most promiscuous compound in our profiling array, with significant interactions to 44 kinases in the panel using a 4°C Tm shift threshold (Fig. 2). More surprising is the broad inhibition profile of the inhibitor Sunitinib (SU11248) and its close analogue SU11652, which have been developed as receptor tyrosine kinase inhibitors and interacted with 25 and 30 Ser/Thr kinases, respectively (>4°C Tm shift). This finding is supported by the study published by Fabian et al. (11). Because Sunitinib is an approved anti-cancer drug, further studies on the cellular consequences of inhibiting these kinases are potentially warranted. Another inhibitor with a very broad inhibition spectrum is Triazolodiamine 1. Close analogues of this inhibitor have been developed as dual Cdk/Aurora inhibitors and are now in clinical trials (19). The identified lack of selectivity by Triazolodiamine 1 raises the possibility that its derivatives also target more kinases in vivo than was originally designed for.
Sensitivity of Ser/Thr Kinases for Inhibition.
Our results show that there are significant differences in “druggability” of the studied kinases using the currently available chemical space. Whereas some targets, such as SLK and ASK1, interact significantly with a variety of ligands, others, like PBK and ERK3, show no binding at all to this set of inhibitors. The resistance of ERK3 to ATP competitive kinase inhibitors can be explained by its structure [Protein Data Bank (PDB) ID code 2I6L], which has a distorted P-loop occupying the ATP-binding pocket. Thus, it is tempting to speculate that inactive kinases that did not interact with inhibitors have their active sites partially blocked. In agreement with that notion we observed that inhibitor sensitivity often correlated with the autophosphorylation status of the kinases (an indication of enzyme activity). Kinases that showed no binding (PBK, VRK1, VRK3, and ERK3) were expressed as nonphosphorylated proteins, and inactive kinases, such as p38δ, ERK1, OSR1, and NDR1, showed low hit rates sometimes restricted to specific inhibitor classes (Fig. 2).
Surprising Selectivity Within Kinase Subfamilies.
A number of kinases in this study cluster in groups of functionally related, structurally related, and sequence-related enzymes. These include the group II PAKs (PAK4, PAK5, and PAK6), the casein kinase γ family (CK1γ1, CK1γ2, and CK1γ3), the CLK family members (CLK1, CLK2, and CLK3), and PIM kinases (PIM1, PIM2, and PIM3). Such clustering is very helpful in assessing the perspectives to achieve selectivity within a group of closely related family members.
Group II PAKs and CK1γ family members have strong sequence and structural homology (>70% sequence identity when comparing the catalytic domains). The family members have similar inhibitor profiles, and design of an inhibitor selective for a particular isoform appears challenging (11). For the CLK subfamily the situation is surprisingly different. For example, the comparison of CLK1 and CLK3 showed a general trend of higher ΔTm values for CLK1 using the same inhibitors. The higher binding affinity of inhibitors to CLK1 also translated into similar differences in enzyme kinetic inhibition data. Among the inhibitors tested, a set of four inhibitors showed significantly stronger inhibition for CLK1 compared with CLK3 (A.N.B., unpublished data), and relatively selective inhibitors for CLK1 could be identified. Comparison of inhibition data for the three closely related PIM kinases revealed another example of isoform selectivity. For example, a series of imidazopyridazines were significantly more potent inhibitors for PIM1 than for the highly related kinase PIM2 (20), and also LY333′531 (ruboxistaurin) was selective for PIM1 (ΔTm = 7.9°C and IC50 = 0.2 μM) as compared with PIM2 (ΔTm = 2.1°C and IC50 > 20 μM). The observed selectivity profiles within these two highly conserved kinase families suggest that dynamic parameters like domain flexibility and plasticity of regulatory elements should be considered in structure-based inhibitor design.
Selectivity/Promiscuity Determinants.
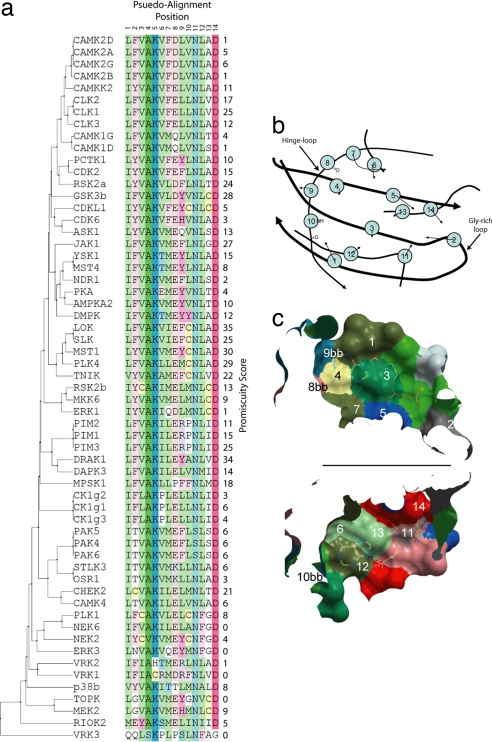
In combination with high-resolution crystal structures, inhibition data provide the opportunity to understand the role of the local sequence and structure toward compound promiscuity. Fig. 3 shows the pseudosequence alignment of the consensus residues that form the ATP-binding site correlated to the promiscuity score (number of hits) for each protein (vide infra). A number of structures exhibit unusual ATP-binding sites that correlate well with low promiscuity scores. For example, CDK6's unusual activation loop blocks the phosphate binding subpocket (PDB ID code 1BI8), and NEK2 shows an unusual activation loop α-helix resulting in the same effect (21). Interestingly, it was observed that there are a number of clusters of proteins with similar ATP-binding site pseudosequences that correlate with high promiscuity scores. The most obvious example of such a cluster is LOK, SLK, MST1, PLK4, and TNIK, whose scores are all equal to or more than 22 hits. The presence of Cys at position 10 in the best-scoring cluster is interesting, but this residue position interacts with the ATP-binding site only via its backbone atoms. It is therefore possible that in this case the change in side-chain size may have an effect on the domain's dynamics along with any change in the actual shape of the active pocket itself. However, a cysteine residue in position 10 is also present in residue combinations with low hit rates (e.g., CDKL1 and NEK2). It is therefore likely that only certain residue compositions and not the presence of a single residue in a fixed position have predictive power determining ligand promiscuity.
Fig. 3.
Alignment and active site. (A) Pseudosequence alignment and phylogenetic tree of the ATP-binding site residues for proteins studied including promiscuity score. (B) A 2D projection of a typical protein kinase ATP-binding site viewed down the axis from the N-terminal lobe through to the C-terminal lobe. The backbone is shown as a black line, which is thickest in the foreground. The residues found to form the ATP-binding site are marked and numbered, and arrows point along the consensus direction of the side chains. “bb” indicates that the residue interacts only via backbone atoms. (C) A cut through the plane of the adenosine ring of ATP of a typical ATP-binding site with ATP included. Upper is viewed from the N-terminal lobe to the C-terminal lobe, and Lower is from the C-terminal lobe up to the N-terminal lobe.
New Applications for Old Drugs?
Food and Drug Administration (FDA)-approved drugs or inhibitors that advanced in clinical testing have been extensively studied and have usually favorable pharmacological properties and safety profiles (22). Unexpected cross-reactivity of such well characterized inhibitors with pharmacologically interesting targets represent therefore particularly exciting hits. A number of well studied inhibitors cross-reacted in our screening panel, and we selected LY333′531 (ruboxistaurin), a PKCβ inhibitor (23) that has been tested in phase III trials for its potential use for diabetic complications. Our screening array revealed a Tm shift of 7.9°C of LY333′531 for PIM1, which translated into an IC50 of 200 nM.
PIM1 is overexpressed in leukemia and other cancer types (24), and the activation of PIM1 plays an important role in malignant transformation by oncogenic tyrosine kinases such as FLT3-ITD, suggesting that PIM inhibition could represent a new therapeutic avenue for hematological malignancies (20, 25). In addition, the primary target of LY333′531, PKCβ, has also been linked to tumor development (26, 27), suggesting that inhibition of both targets may act synergistically on tumor development.
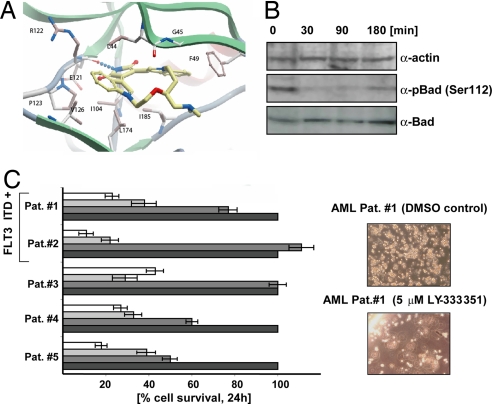
The crystal structure of PIM1 in complex with LY333′531, refined at 1.9-Å resolution, revealed the basis for interaction of this inhibitor with PIM1. LY333′531 binds to the active site of PIM1, forming one hydrogen bond to the hinge region as well as a number of hydrophobic interactions with the active-site residues F49, L120, I104, and I185, showing a remarkable shape complementarity (Fig. 4A).
Fig. 4.
Binding of LY333′531 to PIM1 and effects on primary tumor cells. (A) Active site of PIM1 in complex with LY333′531 solved at 1.9-Å resolution. Hydrogen bonds are indicated by dotted lines, and interacting residues are labeled and shown in ball-and-stick representation. (B) Phosphorylation status of BAD (Ser-112) upon treatment of LY333′531, detected by Western blot analysis. α-Actin levels and total BAD levels are shown as a control. (C) Effects of LY333′531 on the survival of primary tumor cells from five AML patients (Left). A concentration of 0 μM (DMSO), 1 μM, 5 μM, and 10 μM was used, and the increasing concentration of inhibitor has been indicated by black, dark gray, light gray, and white bars. Effect of LY333′531 on clonogenic growth of cells from patient 1 is shown in Right.
LY333′531 effectively induced apoptosis in a dose-dependent manner in human leukemia cell lines that harbor a FLT3-ITD mutation such as MOLM13 (IC50 = 0.7 μM) and MV4;11 (IC50 = 1.5 μM). Short-term exposure of MV4;11 cells to 1 μM LY333′531 led to a significant decrease of phosphorylation of BAD (Fig. 4B), a known PIM1 substrate. In addition, LY333′531 significantly reduced the growth of leukemic blasts from five patients with diagnosed acute myeloid leukemia (AML). As shown in Fig. 4C, exposure of LY333′531 to the leukemic blasts in a short-term liquid culture showed a significant decrease in survival and clonogenic growth in cells from all patients. These preliminary data suggest that LY333′531 and derivatives thereof may be also effective as an anti-tumor agent.
Discussion
Because protein kinases currently constitute a major focus for drug discovery, the availability of systematic interaction data for large arrays of representative chemical inhibitors is highly desirable. Low-molecular-weight inhibitors developed to target the ATP-binding site often display unwanted toxicities in animal models and man, in many cases because of cross-reactivities caused by very similar active-site properties. Ser/Thr kinases play a critical role in cell signaling, and many members of this family have been linked to disease, in particular to cancer (28). However, in drug development Ser/Thr kinases have received less attention than tyrosine kinases, and to date no such inhibitor has been approved for treatment. Because of their implication in many diseases this family may well represent the next generation of targets for kinase drug discovery.
It is highly valuable to obtain information about how amenable the target is for inhibition before embarking on a costly process of developing a clinical candidate. Thus, the striking differences in sensitivity for inhibition reported here show the usefulness of a rapid screening method against a panel of known kinase inhibitors. Even though the size of the used compound set is small compared with a typical high-throughput screening library, it contains a representative and highly diverse set of compounds that have been validated as high-affinity inhibitors for at least one protein kinase. Thus, this study provides a comprehensive guide to the currently pursued kinase inhibitor “chemical space.” Considering the data presented in this study, it comes as no surprise that, despite numerous efforts, no potent and specific ERK1/2 inhibitor has entered clinical trials. Our results show that none of the inhibitor scaffolds in our compound subset are likely to serve as a “platform” for inhibitor development and that, most likely, novel chemical templates would be required to target this kinase. However, targeting upstream or downstream elements of the ERK pathway could provide a valuable alternative. Indeed, MEK1/2 inhibitors have been identified that show promising activity in clinical trials (29).
In contrast, kinases such as MST1, LOK, and DRAK1 are highly sensitive to inhibition. Thus, it is quite likely that many kinase inhibitors targeting the ATP pocket would inhibit one or more of these kinases. Hence, it is of great importance that, in the evaluation of inhibitor specificity, these and targets with similar properties are included in specificity profiling. In a majority of studies on inhibitor specificity, only a small number of kinases are selected, typically those involved in the signal transduction pathway or related family members, which may lead to erroneous conclusions. For example, staurosporine would seem to be a selective kinase inhibitor if tested against the otherwise perfectly reasonable panel consisting of PBK, VRK1, CK1γ, ERK1, ERK3, p38, NEK2, and CDK6.
The high sensitivity of several kinases also provides a compelling argument for the need of more thorough studies of their roles in cellular responses. For example, ASK1 and MST1 are master regulators of apoptosis, and their inhibition is likely to produce unanticipated side effects.
Our pseudoalignment of first-shell active-site residues revealed a correlation of certain residue combinations with ligand promiscuity of the target. However, no single residue position determines this property.
For a number of targets we were not able to identify any inhibitors or identified only weakly binding compounds, and a certain correlation with kinase activity has been noted and has also been discussed before (17). Most kinases exist in both active and inactive form, the latter having a conformation incompatible with either substrate binding or catalysis (30, 31). Some of the clinically successful kinase inhibitors, like imatinib and Sorafenib, target an inactive conformation (32). Because our method of screening does not require the kinase to be in the active form, it can be used to obtain binding profiles to both states. Indeed, we have detected binding of compounds known to bind only to the inactive form as non-ATP-competitive inhibitors. One example for a compound of this class is the specific MEK1/2 inhibitor U0126 (33). In our assay U0126 induced a Tm shift of ≈4°C only with its intended target, MEK2.
The vast majority of the compounds used in this study have emerged from drug discovery programs in the pharmaceutical industry, which suggests that these inhibitors have been optimized for pharmacological and toxicity properties. Most potent in vitro inhibitors fail in preclinical and clinical studies because of absorption, distribution, metabolism, and excretion or toxicological properties as well as a lack of efficacy (34). Clinical inhibitors that failed because of lack of efficacy could thus be excellent candidates for alternative indications. From literature and database searches we concluded that 34 kinases in our screening panel have been linked to cancer, 15 to cardiovascular diseases, and 12 to inflammation-related diseases (SI Table 4). We identified LY333′531 (20) as a potent inhibitor of PIM1 and showed that it could potentially be used for the treatment of leukemia.
In recent years it has become clear that many diseases, most notably cancers, are heterogeneous and adaptable and thus cannot be effectively treated with single agents. The shift toward the development of multitargeting inhibitors is supported by successes of inhibitors such as Sunitinib and Sorafenib (35, 36). The completion of kinase inhibition profiles and the associated evaluation of selectivity and promiscuity determinants of both compounds and ligand binding sites, as provided in this study, thus constitute an important step to select more “optimal” clinical candidates for cancer treatment and for other indications where kinases are likely to contribute to the initiation of and drive the progression of disease.
Methods
Protein Expression and Purification.
All proteins were expressed in Escherichia coli. Detailed protocols for expression and purification of each protein can be downloaded from the Structural Genomics Consortium web site (www.sgc.ox.ac.uk). Either a His6 tag (N- or C-terminal) or a GST fusion system was used to aid purification. Recombinant proteins were >95% pure as judged by SDS/PAGE, and protein identity was confirmed by mass spectrometry.
Thermal Stability Measurements.
Thermal melting experiments were carried out by using a Real-Time PCR Mx3005p machine (Stratagene) according to the protocol described by Vedadi et al. (14).
Compounds.
BIM 1, BIM 8, BIM 9, BIM 10, BIM 11, Chelerythrine, Go 6976, K252c, LY 294002, AG 1295, SU 1498, and staurosporine were purchased from LC Laboratories. Gleevec, Tarceva, and Iressa were purchased from Biaffin. BIM 4, BIM 9, and LY333′531 were purchased from Alexis. Sunitinib and Sorafenib were purchased from Sequoila Research. All other compounds were purchased from Calbiochem (EMD Biosciences). Identities of all compounds used in this study are listed in SI Table 2.
Crystallization.
PIM1 was purified and assayed as described (20). Crystals of the PIM1 LY333′531 complex were obtained at 4°C by using sitting drops mixing 150 nl of PIM1 (10 mg/ml in 50 mM Hepes, pH 7.5/280 mM NaCl/5% glycerol/10 mM DTT) with 50 nl of a solution containing 20% PEG 1K, 0.20 M Li2SO4, 0.1 M citrate/phosphate buffer (pH 4.2). LY333′531 was added to the protein solution with an end concentration of 1 mM from a 50 mM DMSO stock. Diffraction data were collected on a fast-rotating anode model E (FRE) detector equipped with a high-throughput crystallography (HTC) image plate system (Rigaku). Diffraction data were processed, phased, and refined as described (20).
Ranking of Targets by Estimated Promiscuity.
For each protein target the estimated ligand promiscuity was calculated based on the screening results. For each protein a score of 1 was assigned where the tested compound gave a temperature shift of >4 K. The total for each protein was then calculated and normalized by multiplying with the fraction of compounds tested. The final value was multiplied by 100 to provide an easily readable “promiscuity score.”
Core ATP-Binding Pocket Sequence Pseudoalignment.
To obtain an ATP-binding “pocket-centric” structural alignment, four structurally conserved residues that form the top, bottom, and hinge region of the adenosine-binding volume of the ATP-binding set were identified (for example, residues 65, 122, 123, and 174 in the PIM1 structure with PDB ID code 1XWS). Structural superposition was then performed by using ICM (37) on all kinases in our panel for which a structure at a resolution equal to or better than 2.8 Å existed in the PDB, including structures generated in our laboratory (SI Table 1) [CDK2, PDB ID code 1HCK (38); CDK6, PDB ID code 1BI8 (39); CHK2, PDB ID codes 2CN5 and 2CN8 (40); MEK2, PDB ID code 1S9I (41); GSK3β, PDB ID code 1PYX (42)]. From this structural alignment, the residues contacting the cocrystallized compounds in the majority of structures were identified and the consensus sequence positions of these residues were determined. These positions were then identified in a multiple sequence alignment generated by ICM's ZEGA alignment method (37). The pseudoalignment represents an ATP-binding “site-centric” description at the sequence level, thereby removing any noise that is included when considering the whole multiple sequence alignment. The pseudoalignment was then realigned in ICM to reorder and cluster the sequences according to sequence similarity.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Prof. Aled Edwards for critical reading of the manuscript, GlaxoSmithKline for supplying us with the inhibitor VX-680, Dr. F. Niesen for help with screening experiments, and J. F. Debrenzeni for refining the Pim LY333′531 complex. J.S. is supported by the Gertrude von Meissner Foundation. The Structural Genomics Consortium is a registered charity (no. 1097737) that receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research, and the Wellcome Trust.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates of the PIM1-LY333531 complex have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2J2I).
This article contains supporting information online at www.pnas.org/cgi/content/full/0708800104/DC1.
References
- 1.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Cohen P. Nat Rev Drug Discovery. 2002;1:309–315. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- 3.Imming P, Sinning C, Meyer A. Nat Rev Drug Discovery. 2006;5:821–834. doi: 10.1038/nrd2132. [DOI] [PubMed] [Google Scholar]
- 4.Vieth M, Sutherland JJ, Robertson DH, Campbell RM. Drug Discovery Today. 2005;10:839–846. doi: 10.1016/S1359-6446(05)03477-X. [DOI] [PubMed] [Google Scholar]
- 5.Ohno R. Int J Clin Oncol. 2006;11:176–183. doi: 10.1007/s10147-006-0582-5. [DOI] [PubMed] [Google Scholar]
- 6.Bain J, McLauchlan H, Elliott M, Cohen P. Biochem J. 2003;371:199–204. doi: 10.1042/BJ20021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blencke S, Zech B, Engkvist O, Greff Z, Orfi L, Horvath Z, Keri G, Ullrich A, Daub H. Chem Biol. 2004;11:691–701. doi: 10.1016/j.chembiol.2004.02.029. [DOI] [PubMed] [Google Scholar]
- 8.Davies SP, Reddy H, Caivano M, Cohen P. Biochem J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheinerman FB, Giraud E, Laoui A. J Mol Biol. 2005;352:1134–1156. doi: 10.1016/j.jmb.2005.07.074. [DOI] [PubMed] [Google Scholar]
- 10.Vieth M, Higgs RE, Robertson DH, Shapiro M, Gragg EA, Hemmerle H. Biochim Biophys Acta. 2004;1697:243–257. doi: 10.1016/j.bbapap.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 11.Fabian MA, Biggs WH, III, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, et al. Nat Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 12.Bain J, Plater L, Elliott M, Shpiro N, Hastie J, McLauchlan H, Klevernic I, Arthur S, Alessi D, Cohen P. Biochem J. 2007 Sep 13; doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bullock AN, Debreczeni JE, Fedorov OY, Nelson A, Marsden BD, Knapp S. J Med Chem. 2005;48:7604–7614. doi: 10.1021/jm0504858. [DOI] [PubMed] [Google Scholar]
- 14.Vedadi M, Niesen FH, Allali-Hassani A, Fedorov OY, Finerty PJ, Jr, Wasney GA, Yeung R, Arrowsmith C, Ball LJ, Berglund H, et al. Proc Natl Acad Sci USA. 2006;103:15835–15840. doi: 10.1073/pnas.0605224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kervinen J, Ma H, Bayoumy S, Schubert C, Milligan C, Lewandowski F, Moriarty K, Desjarlais RL, Ramachandren K, Wang H, et al. Arch Biochem Biophys. 2006;449:47–56. doi: 10.1016/j.abb.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 16.Kroe RR, Regan J, Proto A, Peet GW, Roy T, Landro LD, Fuschetto NG, Pargellis CA, Ingraham RH. J Med Chem. 2003;46:4669–4675. doi: 10.1021/jm030120s. [DOI] [PubMed] [Google Scholar]
- 17.Mayhood TW, Windsor WT. Anal Biochem. 2005;345:187–197. doi: 10.1016/j.ab.2005.07.032. [DOI] [PubMed] [Google Scholar]
- 18.Bakare O, Ashendel CL, Peng H, Zalkow LH, Burgess EM. Bioorg Med Chem. 2003;11:3165–3170. doi: 10.1016/s0968-0896(03)00267-0. [DOI] [PubMed] [Google Scholar]
- 19.Emanuel S, Rugg CA, Gruninger RH, Lin R, Fuentes-Pesquera A, Connolly PJ, Wetter SK, Hollister B, Kruger WW, Napier C, et al. Cancer Res. 2005;65:9038–9046. doi: 10.1158/0008-5472.CAN-05-0882. [DOI] [PubMed] [Google Scholar]
- 20.Pogacic V, Bullock AN, Fedorov O, Filippakopoulos P, Gasser C, Biondi A, Meyer-Monard S, Knapp S, Schwaller J. Cancer Res. 2007;67:6916–6924. doi: 10.1158/0008-5472.CAN-07-0320. [DOI] [PubMed] [Google Scholar]
- 21.Rellos P, Ivins FJ, Baxter JE, Pike A, Nott TJ, Parkinson DM, Das S, Howell S, Federov O, Shen QY, et al. J Biol Chem. 2007;282:6833–6842. doi: 10.1074/jbc.M609721200. [DOI] [PubMed] [Google Scholar]
- 22.Chong CR, Sullivan DJ., Jr Nature. 2007;448:645–646. doi: 10.1038/448645a. [DOI] [PubMed] [Google Scholar]
- 23.Jirousek MR, Gillig JR, Gonzalez CM, Heath WF, McDonald JH, III, Neel DA, Rito CJ, Singh U, Stramm LE, Melikian-Badalian A, et al. J Med Chem. 1996;39:2664–2671. doi: 10.1021/jm950588y. [DOI] [PubMed] [Google Scholar]
- 24.Roh M, Gary B, Song C, Said-Al-Naief N, Tousson A, Kraft A, Eltoum IE, Abdulkadir SA. Cancer Res. 2003;63:8079–8084. [PubMed] [Google Scholar]
- 25.Adam M, Pogacic V, Bendit M, Chappuis R, Nawijn MC, Duyster J, Fox CJ, Thompson CB, Cools J, Schwaller J. Cancer Res. 2006;66:3828–3835. doi: 10.1158/0008-5472.CAN-05-2309. [DOI] [PubMed] [Google Scholar]
- 26.Koivunen J, Aaltonen V, Peltonen J. Cancer Lett. 2006;235:1–10. doi: 10.1016/j.canlet.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 27.Hans CP, Weisenburger DD, Greiner TC, Chan WC, Aoun P, Cochran GT, Pan Z, Smith LM, Lynch JC, Bociek RG, et al. Mod Pathol. 2005;18:1377–1384. doi: 10.1038/modpathol.3800434. [DOI] [PubMed] [Google Scholar]
- 28.Capra M, Nuciforo PG, Confalonieri S, Quarto M, Bianchi M, Nebuloni M, Boldorini R, Pallotti F, Viale G, Gishizky ML, et al. Cancer Res. 2006;66:8147–8154. doi: 10.1158/0008-5472.CAN-05-3489. [DOI] [PubMed] [Google Scholar]
- 29.Kohno M, Pouyssegur J. Ann Med. 2006;38:200–211. doi: 10.1080/07853890600551037. [DOI] [PubMed] [Google Scholar]
- 30.Adams JA. Chem Rev. 2001;101:2271–2290. doi: 10.1021/cr000230w. [DOI] [PubMed] [Google Scholar]
- 31.Engh RA, Bossemeyer D. Adv Enzyme Regul. 2001;41:121–149. doi: 10.1016/s0065-2571(00)00010-8. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Gray NS. Nat Chem Biol. 2006;2:358–364. doi: 10.1038/nchembio799. [DOI] [PubMed] [Google Scholar]
- 33.Duncia JV, Santella JB, III, Higley CA, Pitts WJ, Wityak J, Frietze WE, Rankin FW, Sun JH, Earl RA, Tabaka AC, et al. Bioorg Med Chem Lett. 1998;8:2839–2844. doi: 10.1016/s0960-894x(98)00522-8. [DOI] [PubMed] [Google Scholar]
- 34.Kola I, Landis J. Nat Rev. 2004;3:711–715. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- 35.Faivre S, Kroemer G, Raymond E. Nat Rev Drug Discovery. 2006;5:671–688. doi: 10.1038/nrd2062. [DOI] [PubMed] [Google Scholar]
- 36.Lierman E, Folens C, Stover EH, Mentens N, Van Miegroet H, Scheers W, Boogaerts M, Vandenberghe P, Marynen P, Cools J. Blood. 2006;108:1374–1376. doi: 10.1182/blood-2006-02-004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abagyan RA, Batalov S. J Mol Biol. 1997;273:355–368. doi: 10.1006/jmbi.1997.1287. [DOI] [PubMed] [Google Scholar]
- 38.Schulze-Gahmen U, Brandsen J, Jones HD, Morgan DO, Meijer L, Vesely J, Kim SH. Proteins. 1995;22:378–391. doi: 10.1002/prot.340220408. [DOI] [PubMed] [Google Scholar]
- 39.Russo AA, Tong L, Lee JO, Jeffrey PD, Pavletich NP. Nature. 1998;395:237–243. doi: 10.1038/26155. [DOI] [PubMed] [Google Scholar]
- 40.Oliver AW, Paul A, Boxall KJ, Barrie SE, Aherne GW, Garrett MD, Mittnacht S, Pearl LH. EMBO J. 2006;25:3179–3190. doi: 10.1038/sj.emboj.7601209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohren JF, Chen H, Pavlovsky A, Whitehead C, Zhang E, Kuffa P, Yan C, McConnell P, Spessard C, Banotai C, et al. Nat Struct Mol Biol. 2004;11:1192–1197. doi: 10.1038/nsmb859. [DOI] [PubMed] [Google Scholar]
- 42.Bertrand JA, Thieffine S, Vulpetti A, Cristiani C, Valsasina B, Knapp S, Kalisz HM, Flocco M. J Mol Biol. 2003;333:393–407. doi: 10.1016/j.jmb.2003.08.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.