Abstract
Culture of bronchoalveolar lavage fluid (BALF) is the gold standard for detection of pathogens in the lower airways in cystic fibrosis (CF). However, current culture results do not explain all clinical observations in CF, including negative culture results during pulmonary exacerbation and inflammation in the absence of pathogens. We hypothesize that organisms not routinely identified by culture occur in the CF airway and may contribute to disease. To test this hypothesis we used a culture-independent molecular approach, based on use of rRNA sequence analysis, to assess the bacterial composition of BALF from children with CF and disease controls (DC). Specimens from 42 subjects (28 CF) were examined, and ≈6,600 total clones were screened to identify 121 species of bacteria. In general, a single rRNA type dominated clone libraries from CF specimens, but not DC. Thirteen CF subjects contained bacteria that are not routinely assessed by culture. In four CF subjects, candidate pathogens were identified and include the anaerobe Prevotella denticola, a Lysobacter sp., and members of the Rickettsiales. The presumptive pathogens Tropheryma whipplei and Granulicatella elegans were identified in cases from the DC group. The presence of unexpected bacteria in CF may explain inflammation without documented pathogens and consequent failure to respond to standard treatment. These results show that molecular techniques provide a broader perspective on airway bacteria than do routine clinical cultures and thus can identify targets for further clinical evaluation.
Keywords: pulmonary microbiology, ribosomal RNA
Cystic fibrosis (CF), the most common autosomal lethal disease in Caucasians, is caused by mutation in the CF transmembrane conductance regulator gene, which results in a generalized exocrinopathy (1). Thickened secretions caused by improper regulation of airway surface liquid contribute to the accumulation of mucus in the airway and defective mucociliary clearance (2). Retained mucus plugs provide a niche for bacterial colonization and persistence. Most morbidity and mortality associated with CF is attributed to microbial infections in the airway and the persistent inflammatory response (3).
A small number of pathogens are recognized traditionally in CF airway disease. These include Pseudomonas aeruginosa, Staphylococcus aureus, Haemophilus influenzae, and the Burkholderia cepacia complex (4–7). Other bacteria, including Stenotrophomonas maltophilia and Achromobacter xylosoxidans, also are associated with CF, but there is controversy as to their roles as pathogens (8–10). This view of bacteria associated with CF is based on culture, however, which may not recover or identify all bacteria present in a specimen. Although known pathogens clearly are important in airway disease, a more comprehensive picture of the bacterial community in the CF airway potentially could lead to improved insight about the disease and thus better treatment. For instance, “normal” microbiota that are commonly present, but not considered pathogenic, may affect the disease course (11). In addition, there are documented cases of inflammation with no cultured pathogen (12–14).
Developments in environmental microbiology have obviated, to considerable extent, the requirement to culture microbes to detect and identify them (15). Instead of culture tests, rRNA genes are harvested by PCR directly from samples, and their sequences are used to identify bacteria in the sample. In the current study, we have used this culture-independent molecular technique to determine the composition of bacterial communities associated with bronchoalveolar lavage fluid (BALF) from CF and disease control (DC) subjects. We compare the results to those obtained by culture and conclude that molecular analysis provides a more comprehensive view of the microbiology of the lower airway than is obtained by standard laboratory culture.
Results
Molecular Summary.
A total of 69 rRNA gene libraries (50 CF) were prepared with bacteria-specific PCR amplicons obtained from 42 subjects (28 CF; Fig. 1).Replicate libraries from the same sample were constructed from five CF subjects to examine the reproducibility of the procedures, and results were highly similar in all cases. Libraries prepared from three CF subjects and two DC subjects contained sequences of uncertain source, more similar to water samples (e.g., content of Methylobacterium spp.) than to other clinical samples and were excluded from the study. Overall, ≈6,600 clones were screened by restriction fragment length polymorphism (RFLP) to identify unique types for sequence analysis and btain a census of the different sequence types in each library. Approximately ≈1,600 unique sequences were determined, each ≈800 nt in length. Sequences identified 121 bacterial species (65 in CF) with 33 species observed in both groups.
Fig. 1.
Flow chart of specimens and PCR results for CF and DC specimens. Not all specimens were positive for PCR. For each specimen one to three processing types were examined. In 22 cases multiple libraries were constructed from the same specimen. Fifteen specimens were positive for PCR with both the hard- and soft-spin pellets and are labeled P1/P2. Replicate libraries were made in the remaining seven cases and are labeled replicates. Specimens with a single library constructed are labeled single.
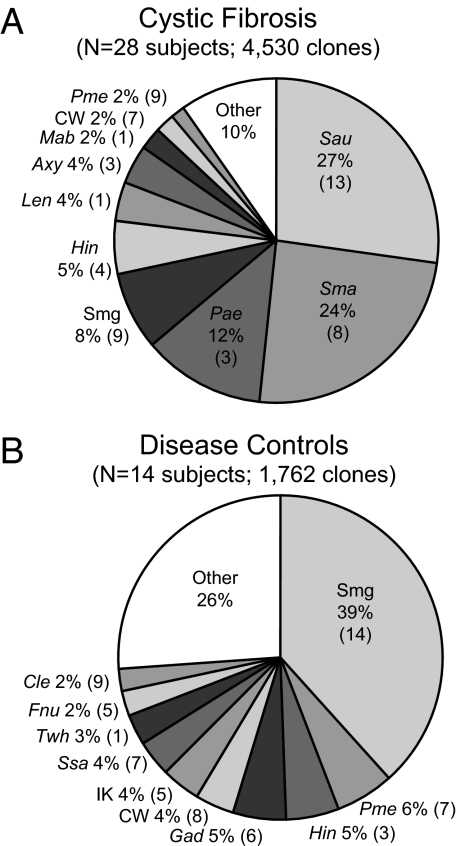
The molecular microbiology among the specimen sets was complex in makeup, so we grouped the results according to the similarities of the sets by using UniFrac. Results are summarized in Fig. 2. The overall diversity observed in CF was distinct from that seen in the DC (Fig. 2). The most frequently encountered sequences in CF were members of relatively few species, generally less complex than clone libraries from DC subjects [summarized in Fig. 3 and supporting information (SI) Table 1].
Fig. 2.
Bacterial community comparisons of BALF-derived PCR libraries were determined with weighted UniFrac, and the relationships are displayed by the unweighted pair group method with arithmetic mean clustering algorithm. The numbers in the dendogram represent the jackknife support for the labeled nodes. Cohort and patient numbers were used to label each subject. Asterisks are used to mark CF subjects with samples that did not yield any pathogenic isolates. The bars represent the percentage of clones indicative of aerobic organisms (open) and anaerobic organisms (filled). The number of samples, specimens, clones identified by sequence (including RFLP counts), organisms, and prevalence of the most common organism identified are also given. The tree is labeled by phylogenetic identity (genus and division) of the most common organism identified in each subject.
Fig. 3.
Distribution of the 10 most prevalent organisms observed by subject groups. (A) Summary of the CF cohort. (B) The same comparisons as in A for the DC cohort. Organism names are abbreviated as follows: Sau, S. aureus; Sma, S. maltophilia; Pae, P. aeruginosa; Smg, Streptococcus mitis group; Hin, H. influenzae; Len, Lysobacter enzymogenes; Axy, A. xylosoxidans; Mab, Mycobacterium abscessus; CW, Porphyromonas sp. CW034;, Pme, Prevotella melaninogenica; Gad, Granulicatella adiacens; IK, Prevotella sp. IK062; Ssa; Streptococcus sanguis; Twh, Tropheryma whipplei; Fnu, Fusobacterium nucleatum; Cle, Capnocytophaga leadbetteri. The numbers in parentheses give the numbers of subjects in which each organism was observed.
Libraries from CF Subjects.
Although variable in composition between subjects, libraries from CF subjects generally were dominated (>85% of total) by sequences characteristic of known CF pathogens (17/28, 61%). About half of the 28 CF subjects contained a single predominant pathogen (>85% of library; Fig. 1). Two CF pathogens dominated two additional subjects. The remaining subjects (11/28, 39%) yielded sequences indicative of organisms not routinely identified in the airway of subjects with CF. Five of these 11 (55%) were scored negative for pathogens by culture (denoted by * in Fig. 2).
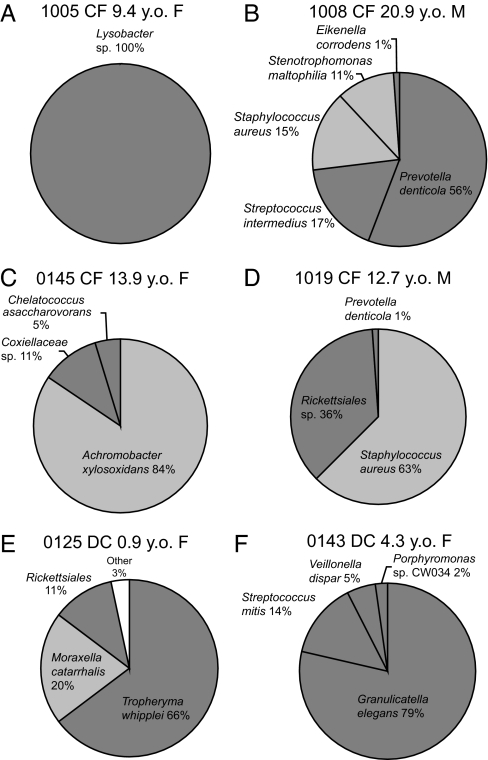
Four specimens contained sequences that likely represent novel pathogens in the CF airway. One candidate for a new CF pathogen is a member of the genus Lysobacter, which is not usually considered a pathogen but is closely related (97% rRNA sequence identity) to pathogenic members of the genus Stenotrophomonas. Only sequences related to Lysobacter were recovered from the specimen from subject 1005 (Fig. 4A). Culture failed to isolate any organisms from the specimen, but Gram-negative rods were observed.
Fig. 4.
Pie charts of the distribution of sequences from selected libraries that contained unexpected sequences. (A–D) Subjects with CF. (E and F) DC subjects. Dark gray portions represent sequences that were not expected based on traditional culture results. Light gray portions represent sequences representative of pathogens detected by both molecular and culture methods.
BALF from subject 1008 yielded four potential pathogens, only two of which were detected by culture, S. aureus and S. maltophilia; the others were a dominant Prevotella denticola and Streptococcus intermedius (Fig. 4B). Results from this specimen illustrate the capacity of the molecular technology to readily detect polymicrobial disease and potentially contribute to clinical evaluation.
Two CF specimens contained sequences that represent novel examples of α-Proteobacteria specifically related to members of the Coxiellaceae (Fig. 4C) and Rickettsiales (Fig. 4D). In both cases these sequences occurred with those of known CF pathogens. However, members of both the Coxiellaceae and Rickettsiales are typically considered pathogens and usually occupy an intracellular environment in host cells. Such organisms are not detected by routine culture and, considering the abundance of such organisms in patient samples, should be considered of clinical concern.
In most CF cases in this study typical CF pathogens were identified. However, in 25% of the CF subjects examined the microbial communities did not contain typical CF pathogens. The clinical relevance of the organisms observed is not clear, but the relative abundance of anaerobic bacteria in these libraries was higher (Fig. 2). These subjects contained predicted anaerobic bacteria in the range of 27% to 93% of the clones examined. Such anaerobic bacteria were rare in CF subjects with typical pathogens.
DC Libraries.
Only 4/14 of the DC libraries contained a relatively low complexity of sequences (≤5 species); other DC specimens contained higher complexity (Fig. 2). Low-complexity libraries reflect enrichment of particular organisms and so possibly signify bacterial involvement in disease. The high-complexity libraries may have clinical involvement, but establishing that would require a larger study to assure higher statistical power. We do not consider the high-complexity libraries further.
Two of the low-complexity DC libraries contained a single sequence type, in one case Streptococcus mitis group (which includes S. pneumoniae) sequences and in the other H. influenzae sequences. The organisms observed are documented human pathogens in the airway and in both cases culture results (reported as “alpha strep not S. pneumoniae” and “H. influenzae,” respectively) were consistent with molecular results.
The remaining two subjects with low-complexity libraries from the DC contained unexpected sequences that may indicate microbial disease. In one subject the majority of the sequences from the library (66%) were indicative of Tropheryma whipplei, the causative agent of Whipple's disease. In addition, Moraxella cattharalis and a novel member of the Rickettsiales (95% identification with sequences in Fig. 4D) were present in the library (Fig. 4E). The second subject contained sequences related to Granulicatella elegans and Streptococcus mitis group (Fig. 4F).
Comparison to Culture.
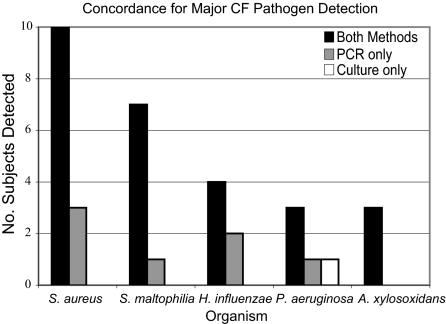
Clinical culture results were compared with molecular results and are summarized in Fig. 5. Only 3 of the 14 DC subjects were culture positive for recognized CF bacterial pathogens. In contrast, 22 (79%) of the CF specimens were culture positive for such organisms. The molecular survey detected the cultured CF pathogen in 27 of 28 subjects (96%). In seven subjects (25%) the rDNA sequence analysis, but not culture, detected CF pathogens (Fig. 5). The relatively low numbers of subjects with P. aeruginosa was of note. We looked at the culture history in the year before bronchoscopy to determine why the incidence was lower than expected in these subjects. P. aeruginosa was detected in 12/28 subjects (43%) in the prior year (SI Table 2).
Fig. 5.
Summary of major CF pathogens detected by both culture and molecular analysis with all-bacteria primers. Dark bars represent detection by both methods, gray bars represent detection by molecular methods only, and empty bars represent detection by culture only.
Discussion
This and previous studies show that the sequence-based molecular approach to clinical microbiology provides microbiological perspective that can significantly enhance the standard clinical culture-based view (16–19). The use of sequences for identification in this study is a substantial improvement over electrophoretic patterns for the identification of microbes, because there is no ambiguity in identity. The two patient cohorts examined, CF and DC, were not age-matched, which may limit some comparisons between the two cohorts. Moreover, we acknowledge that the CF cohort is small and certainly does not represent the full spectrum of the disease. Nevertheless, this study is to date the most extensive sequence-based analysis of bacteria associated with the human airway to our knowledge.
Molecular results with most specimens matched the culture results with regard to conventionally recognized pathogens. In addition, several specimens from CF contained sequences that were unexpected and possibly identify new pathogens. In other CF specimens, likely pathogens (P. denticola, Coxiellaceae and Rickettsiales; Fig. 4) were observed along with known pathogens. Expanded studies will be required to test the specific association of these potentially novel pathogens with CF and determine clinical manifestations.
One technical issue of concern in sampling the airway is the possibility of oral or upper respiratory contamination of the specimen as the bronchoscope is advanced into the lung. The sequence-based methods used here, with general bacterial PCR primers, sample the most abundant organisms present in specimens. Oral and upper airway suites of microbes are more complex and generally different phylogenetically from those we encountered in BALF specimens (20). We cannot rule out low-incidence sequences as upper airway contaminants, but the types and relatively low complexity of sequences in these BALF specimens both support their specificity to the lower airway.
Whether or not there are clinical consequences of anaerobic bacteria in the airways of CF subjects is an open question. It is not clear whether BAL consistently samples mucus from the airways; in many cases, the BALF remains clear without any significant mucus plugs present. Thus, anaerobic bacteria, which are expected to occur mainly in mucus plugs, may often be present but not sampled. There were no distinguishing features among the CF subjects in this study that contained high levels of anaerobic bacteria. A larger study will be required to elucidate any clinical features that predict involvement of anaerobic bacteria in exacerbation.
In addition to the results from CF specimens, several bacteria with potential clinical significance were detected in DC. In the single case of a lung abscess analyzed, culture had failed to identify any bacteria, but the molecular method detected Granulicatella elegans as 79% of the clones in the library prepared from the specimen (Fig. 4F). This organism previously has been associated with endocarditis (21). In a second case, diagnosed as pneumonia, culture results reported the presence of α-hemolytic Streptococcus (not S. pneumoniae). The sequence from this specimen, however, shared 99.5% sequence identity with those obtained from a CF specimen where S. pneumoniae was identified. This result demonstrates the utility of the molecular perspective in identification of organisms even in the context of uncertainty in culture-based identification. The result also demonstrates the utility of the molecular methods to address bacteria associated with pulmonary disease other than CF.
Tropheryma whipplei, the causative agent of Whipple's disease (22), was the most frequently encountered sequence in one of the DC specimens from an interstitial lung disease (ILD) subject. In addition, sequences belonging to the Rickettsiales group were present at lower levels in this specimen. Neither of these organisms is detected by standard culture. It is possible that the occurrence of one or both organisms may correlate with a particular subset of ILD patients and thereby provide diagnostic information relevant to therapy.
rRNA sequences obtained from environmental (including clinical) samples seldom are identical to previously encountered sequences. In this study, sequences were assigned to named taxa based on sequence similarities (approximately ≥97% sequence identity). Although organisms with similar sequences are expected to share many physiological properties, we note that small differences in rRNA sequences may not reflect genomic-level differences with significant clinical considerations, for instance drug resistance vs. sensitivity. The primary goal of rRNA sequence analysis is to identify bacteria in the clinical specimens, and it does so unequivocally. Determination of physiological properties such as antibiotic resistance requires information beyond the specificity of rRNA sequences, either molecular- or culture-based results.
Methods based on bacterial gene sequences can complement standard clinical microbiology for characterization of bacteria associated with disease states and are increasingly economical. rRNA gene sequences are the most widely used molecular measure for bacterial identification. Consequently, an rRNA sequence represents an important first evaluation of an organism, its basic nature at the species level. Based on that knowledge, additional genes, for instance for pathogenic factors, can be analyzed. Thus, culture-independent molecular approaches can provide new clinical tools for the detection, identification, and understanding of bacterial interactions in disease processes. Detection of unexpected bacteria may explain failure to respond to standard treatment and thereby identify new targets for antibiotic therapies in CF.
Materials and Methods
Subjects and Specimens.
All human material was obtained under Colorado Combined Institutional Review Board-approved protocols at The Children's Hospital in Denver. Informed consent was obtained from all subjects or their legal guardians. The 36 CF subjects comprised 19 females with a median age of 9.1 years (range 1.0 to 20.5) and 17 males with median age 8.5 years (range 0.9 to 20.9). The DC contained 11 females with median age 3.5 years (range 0.5 to 9.1) and 10 males with median age 2.7 years (range 0.7 to 17.7). Eighteen CF subjects were homozygous ΔF508, 12 were heterozygous ΔF508 with various other mutant alleles, a single CF subject was G542X/unknown at the CFTR locus, and no genotype was available for five subjects.
Up to 400 μl (range 10 to 400 μl) of BALF (n = 35, 29 CF), or 50 μl of pelleted material from BALF [n = 33 (15 CF) P1 pellets (750 [times) g) and n = 31 (14 CF) P2 pellets (4,000 × g)] was extracted by using a modified bead beating and solvent extraction protocol (23). The DNA was precipitated, air-dried, and resuspended in 50 μl of TE [10 mM Tris (pH 8.0), 1 mM EDTA]. For P2 pellet specimens 15/31 (48%) yielded measurable DNA after extraction. All extractions included a negative extraction control where buffer only was extracted in parallel with specimens. Culture microbiology was performed in the clinical microbiology lab at The Children's Hospital by using standard techniques developed for CF (24).
rRNA gene sequences were amplified in 50 μl of PCRs, which included (final concentrations): 1–50 ng of DNA in reaction mixtures, 1× Hotmaster TaqPCR buffer (Eppendorf), 2.5 mM MgCl2, 100 μM each deoxynucleoside triphosphate, 300 μM each forward (27F) and reverse (907R) primer (25, 26), and 1.25 units of Hotmaster Taq (Eppendorf). Reaction mixtures were incubated in a PTC-100 Programmable Thermal Cycler (MJ Research) using a touch-down PCR (TD-PCR) protocol with a 2-min initial denaturation step at 94°C followed by 20 cycles at 92°C for 15 s, 65°C annealing for 15 s, and 65°C extension for 1 min. The annealing temperature was decreased by 1° each cycle. Finally, 11–15 cycles of PCR were performed by using the same cycles as above, but with 45°C annealing in all steps. A final extension step of 20 min at 65°C was performed to ensure all products contained intact ends for cloning. To determine whether any PCR inhibitors were present, poisoning controls were performed, by spiking purified Escherichia coli genomic DNA into replicate PCRs. In cases where no rDNA PCR product was evident, human β-actin controls were performed as in ref. 27.
PCR products were cloned into the TOPO TA cloning kit vector (Invitrogen) as specified by the manufacturer. Plasmid DNAs (96 per library) containing unique inserts were sorted by RFLP analysis (23). Plasmid DNA was used as template for PCR (30 cycles, 92°C for 15 s, 52°C for 15 s, 72°C for 1 min) with vector primers T3 and T7. Sequence information was gathered by using either a LICOR 4200 or Molecular Dynamics MegaBACE 1000 DNA sequencer with vector primers T3 and T7.
Sequence Analysis.
Contiguous sequences from multiple sequencing reactions were assembled by using the in-house program Xplorseq 2.0 (Daniel Frank, personal communication) and compared with the GenBank database by using BLAST running locally to determine their approximate phylogenetic relationships (28). Sequences were aligned by using NAST (29) and inserted into the ARB software package (30). Each sequence was then assigned an organism name based on the phylogenetic placement in ARB. Chimeras were detected by comparing the best match by BLAST for each end of the sequence. Sequences that were considered chimeric were excluded. Community comparisons were made by using weighted Unifrac (31, 32) with the ARB parsimony insertion tree.
Supplementary Material
ACKNOWLEDGMENTS.
This work was supported by the University of Colorado Butcher Genomics-Biotechnology Initiative, Cystic Fibrosis Foundation Grant PACE04A0, the General Clinical Research Program, the National Center for Research Resources, National Institutes of Health Grant M01 RR00069, and Clinical Proteomics Center in Childhood Lung Disease Grant U01 HL081335.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY805987–AY806453, DQ188268–DQ188805, and EU111806–EU112454).
This article contains supporting information online at www.pnas.org/cgi/content/full/0709804104/DC1.
References
- 1.Welsh M, Ramsey B, Accurso F, Cutting G. In: The Metabolic and Molecular Basis of Inherited Diseases. Scriver C, Beaudet A, Sly W, Valle D, editors. New York: McGraw–Hill; 2001. pp. 5121–5188. [Google Scholar]
- 2.Matsui H, Randell SH, Peretti SW, Davis CW, Boucher RC. J Clin Invest. 1998;102:1125–1131. doi: 10.1172/JCI2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brennan AL, Geddes DM. Curr Opin Infect Dis. 2002;15:175–182. doi: 10.1097/00001432-200204000-00013. [DOI] [PubMed] [Google Scholar]
- 4.Burns JL, Emerson J, Stapp JR, Yim DL, Krzewinski J, Louden L, Ramsey BW, Clausen CR. Clin Infect Dis. 1998;27:158–163. doi: 10.1086/514631. [DOI] [PubMed] [Google Scholar]
- 5.Gibson RL, Burns JL, Ramsey BW. Am J Respir Crit Care Med. 2003;168:918–951. doi: 10.1164/rccm.200304-505SO. [DOI] [PubMed] [Google Scholar]
- 6.Gilligan PH. Clin Microbiol Rev. 1991;4:35–51. doi: 10.1128/cmr.4.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shreve MR, Butler S, Kaplowitz HJ, Rabin HR, Stokes D, Light M, Regelmann WE. Investigators for the Epidemiologic Study of Cystic Fibrosis. J Clin Microbiol. 1999;37:753–757. doi: 10.1128/jcm.37.3.753-757.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goss CH, Otto K, Aitken ML, Rubenfeld GD. Am J Respir Crit Care Med. 2002;166:356–361. doi: 10.1164/rccm.2109078. [DOI] [PubMed] [Google Scholar]
- 9.Goss CH, Mayer-Hamblett N, Aitken ML, Rubenfeld GD, Ramsey BW. Thorax. 2004;59:955–959. doi: 10.1136/thx.2003.017707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan K, Conway SP, Brownlee KG, Etherington C, Peckham DG. Pediatr Pulmonol. 2002;34:101–104. doi: 10.1002/ppul.10143. [DOI] [PubMed] [Google Scholar]
- 11.Duan K, Dammel C, Stein J, Rabin H, Surette MG. Mol Microbiol. 2003;50:1477–1491. doi: 10.1046/j.1365-2958.2003.03803.x. [DOI] [PubMed] [Google Scholar]
- 12.Armstrong DS, Grimwood K, Carlin JB, Carzino R, Gutierrez JP, Hull J, Olinsky A, Phelan EM, Robertson CF, Phelan PD. Am J Respir Crit Care Med. 1997;156:1197–1204. doi: 10.1164/ajrccm.156.4.96-11058. [DOI] [PubMed] [Google Scholar]
- 13.Balough K, McCubbin M, Weinberger M, Smits W, Ahrens R, Fick R. Pediatr Pulmonol. 1995;20:63–70. doi: 10.1002/ppul.1950200203. [DOI] [PubMed] [Google Scholar]
- 14.Nixon GM, Armstrong DS, Carzino R, Carlin JB, Olinsky A, Robertson CF, Grimwood K, Wainwright C. Arch Dis Child. 2002;87:306–311. doi: 10.1136/adc.87.4.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pace NR. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 16.van Belkum A, Renders NH, Smith S, Overbeek SE, Verbrugh HA. FEMS Immunol Med Microbiol. 2000;27:51–57. doi: 10.1111/j.1574-695X.2000.tb01411.x. [DOI] [PubMed] [Google Scholar]
- 17.Rogers GB, Carroll MP, Serisier DJ, Hockey PM, Jones G, Bruce KD. J Clin Microbiol. 2004;42:5176–5183. doi: 10.1128/JCM.42.11.5176-5183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rogers GB, Carroll MP, Serisier DJ, Hockey PM, Kehagia V, Jones GR, Bruce KD. Respir Res. 2005;6:49–60. doi: 10.1186/1465-9921-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogers GB, Hart CA, Mason JR, Hughes M, Walshaw MJ, Bruce KD. J Clin Microbiol. 2003;41:3548–3558. doi: 10.1128/JCM.41.8.3548-3558.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. J Bacteriol. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohara-Nemoto Y, Kishi K, Satho M, Tajika S, Sasaki M, Namioka A, Kimura S. J Clin Microbiol. 2005;43:1405–1407. doi: 10.1128/JCM.43.3.1405-1407.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Relman D, Schmidt T, MacDermott R, Falkow S. N Engl J Med. 1992;327:293–301. doi: 10.1056/NEJM199207303270501. [DOI] [PubMed] [Google Scholar]
- 23.Dojka MA, Hugenholtz P, Haack SK, Pace NR. Appl Environ Microbiol. 1998;64:3869–3877. doi: 10.1128/aem.64.10.3869-3877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cystic Fibrosis Foundation. Microbiology and Infectious Disease in Cystic Fibrosis; Consensus Conference: Concepts in Care. Vol 5. Bethesda: Cystic Fibrosis Foundation; 1994. pp. 1–26. section 1. [Google Scholar]
- 25.Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lane DJ. In: Nucleic Acid Techniques in Bacterial Systematics. Stackebrandt E, Goodfellow M, editors. New York: Wiley; 1991. pp. 115–175. [Google Scholar]
- 27.Drake WP, Pei Z, Pride DT, Collins RD, Cover TL, Blaser MJ. Emerg Infect Dis. 2002;8:1334–1341. doi: 10.3201/eid0811.020318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeSantis TZ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, Phan R, Andersen GL. Nucleic Acids Res. 2006;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, et al. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lozupone CA, Knight R. Appl Env Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lozupone CA, Hamady M, Kelley ST, Knight R. Appl Env Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.