Abstract
Two lineages of viral RNA-dependent RNA polymerases (RDRPs) differing in the organization (canonical vs. noncanonical) of the palm subdomain have been identified. Phylogenetic analyses indicate that both lineages diverged at a very early stage of the evolution of the enzyme [Gorbalenya AE, Pringle FM, Zeddam JL, Luke BT, Cameron CE, Kalmakoff J, Hanzlik TN, Gordon KH, Ward VK (2002) J Mol Biol 324:47–62]. Here, we report the x-ray structure of a noncanonical birnaviral RDRP, named VP1, in its free form, bound to Mg2+ ions, and bound to a peptide representing the polymerase-binding motif of the regulatory viral protein VP3. The structure of VP1 reveals that the noncanonical connectivity of the palm subdomain maintains the geometry of the catalytic residues found in canonical polymerases but results in a partial blocking of the active site cavity. The VP1–VP3 peptide complex shows a mode of polymerase activation in which VP3 binding promotes a conformational change that removes the steric blockade of the VP1 active site, facilitating the accommodation of the template and incoming nucleotides for catalysis. The striking structural similarities between birnavirus (dsRNA) and the positive-stranded RNA picornavirus and calicivirus RDRPs provide evidence supporting the existence of functional and evolutionary relationships between these two virus groups.
Keywords: birnavirus replication, infectious bursal disease virus, noncanonical palm, virus evolution, double-stranded RNA viruses
Palm-based nucleotide polymerases comprise the largest family of template-dependent polynucleotide polymerases. The palm subdomain, considered a vestige of the ancestral protein polymerase (1), is found in polymerases from all extant RNA and retroid (DNA and RNA) viruses. Within viral RNA-dependent RNA polymerases (RDRPs), two different lineages have been identified differing in a sequence permutation of the palm subdomain. Both lineages diverged at a very early stage of evolution of the enzyme as indicated by phylogenetic analyses (2). Permuted RDRPs belong to an ancient lineage, exclusively found in the dsRNA birnaviruses and in two members of the Tetraviridae family having a positive-stranded RNA genome.
Infectious bursal disease virus (IBDV), an immunosuppressive avian pathogen (3), is the best characterized member of the Birnaviridae family. This family groups single-shelled icosahedral (triangulation number T = 13) viruses with dsRNA bipartite genomes (4). Birnaviruses exhibit a series of structural and biological features, the most conspicuous one being the lack of an internal transcriptional core (5), that clearly differentiate them from the rest of the dsRNA viruses.
The IBDV RDRP (97.5 kDa) is encoded by the smaller genomic segment (segment B) (6). The RDRP forms part of a ribonucleoprotein complex that also includes the scaffolding polypeptide (VP3) and the virus genome (7). Although a fraction of the RDRP molecules are covalently linked to the 5′ end of the dsRNA virus genome segments (8), its encapsidation requires the formation of a complex with VP3 (9).
The structure of the IBDV RDRP, unbound form, has been recently determined (10). Here, we report the crystal structures of the IBDV polymerase in its apo form and bound to Mg2+ ions and to a peptide, containing the polymerase binding motif of protein VP3 (11), at resolutions of 2.4 and 3.1 and 2.7 Å, respectively. The comparison of the different structures determined reveals that the permuted topology of the VP1 palm results in unique structural properties that have consequences in the regulation of the enzyme activity.
Results and Discussion
Overall Structure of IBDV VP1 and Comparison with Other RDRPs.
The structure of a recombinant version of the infectious bursal disease virus RDRP, VP1 (residues 1–845), was determined by combining phases from multiwavelength anomalous dispersion of a Samarium derivative at 3.8-Å resolution and molecular replacement, using native data to 2.4 Å [see Methods and supporting information (SI) Table 1].
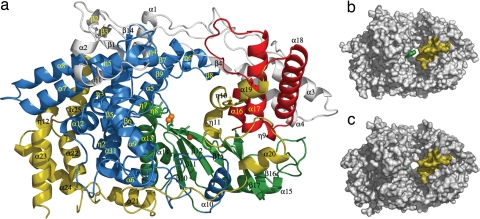
VP1 folds into a compact unit with a roughly oval shape (dimensions 95 × 55 × 54 Å3). The polypeptide chain is organized in 37 helices (25 α-helices and 12 helices 310) and 17 β-strands, and can be divided into three domains by comparison with other viral RDRPs (12): (i) the central polymerase domain (residues 169–657) containing the classical “fingers,” “palm” and “thumb” subdomains found in all other polynucleotide polymerases (13), (ii) an N-terminal domain (residues 1–168), surrounding the central polymerase domain, which bridges fingers and thumb on one side of the catalytic cleft, and (iii) a horseshoe shaped C-terminal domain (residues 658–845) extending through palm and fingers on the other side (Fig. 1).
Fig. 1.
Structure of VP1 polymerase of IBDV. (a) View of a ribbon diagram showing the secondary structural elements explicitly labeled as follows: the N-terminal domain (gray) is folded in five helices (α1–α4 and the 310 helix η1), four β-strands (β1–β4), and three long loops. The VP1 fingers (blue) are organized in two regions. The first region is the inner fingers, consisting primarily of helices α5, α6, α9, α10, α11, and α12 surrounding and packed against the palm subdomain. The second region is the outer fingers, which include the following: (i) a five-stranded β-sheet (β1, β7, β9, β13, β14), with β1 contributed by the N-terminal domain, surrounded by a surface of exposed helices (η4, η5, α7, and α8); (ii) an adjacent region formed by helix η3 and the β-hairpin β5–β6 pointing out of the central cavity of the polymerase; and (iii) a loop formed by β8 and η6 (residues 321–335), which effectively extends toward the thumb domain. The palm subdomain (green) consists of a four-stranded β-sheet (β10, β11, β12, and β15) flanked by two α-helices (α13 and α14) and followed by an additional helix α15 and a long loop connecting the β16–β17 hairpin. The thumb subdomain (red) includes three α-helices (α16, α17, and α18) connected by long loops. Finally, the C-terminal domain (yellow; residues 671–804) includes four helices (α19, η10, η11, and α20), next to the thumb, followed by six α-helices (α21–α25 and η12), close to fingers that are connected by a long and flexible loop. (b) Surface representation of the IBDV VP1 in its apo form, showing the central cleft, which is partially covered by two protrusions that project from the B loop (green) and the α19-η10-η11 region (yellow) at the C-terminal domain. (c) Surface representation of the IBDV VP1 when complexed with the VP3 derived peptide.
The first 26 residues of VP1 are disordered and not visible in the structure. Residues 26–32 point out of the polymerase core and participate in crystal packing interactions. The remainder of the domain, residues 33–168, is organized in four α-helices (α1 to α4), one 310 helix (η1), four β-strands (β1 to β4), and three long loops, covering one side of the active site cleft. Extensive interactions between the N-terminal helices α3 and α4 and helices α17 and α18 of the thumb subdomain are critical for the close conformation of the VP1 polymerase (Fig. 1a).
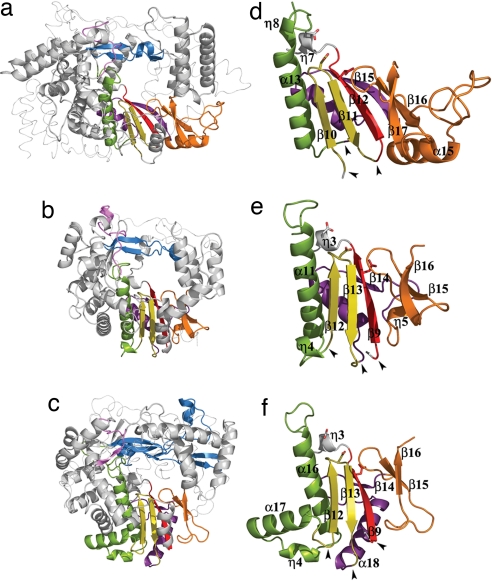
The central polymerase domain of VP1 shows all structural motifs characteristic of RNA-dependent RNA polymerases, involved in the following: nucleotide recognition and binding (A, B, and F), phosphoryl transfer (A and C), structural integrity of the palm (D), priming nucleotide binding (E), and template binding (B, F, and G) (Fig. 2 and SI Fig. 5). The overall architecture of this domain is strikingly similar to those of positive-strand RNA picorna- and caliciviruses (12, 14, 15) (SI Table 2 and Fig. 2).
Fig. 2.
The conserved structural polymerase motifs. (a–c) The polymerase core of IBDV VP1 (a) compared with the equivalent cores in FMDV (b) and bacteriophage φ6 (c) RDRPs. The secondary structural elements containing the conserved motifs are colored as follows: A, red; B, green; C, yellow; D, purple; E, orange; F, blue; G, pink. The N- and C-terminal domains are shown as thin ribbons. (d–e) Structural organization of the permuted palm subdomain of IBDV VP1 (d) compared with the canonical palm subdomains of FMDV (e) and φ6 (f) RDRPs. Side chains of the acidic residues in the polymerase catalytic sites are shown as sticks. The black arrowheads indicate positions where the backbone connectivity is broken in the canonical polymerases and reconnected in the permuted IBDV RDRP.
The C-terminal domain of IBDV VP1 includes helices α19, η10, η11, and α20, close to the thumb subdomain, followed by five α-helices (α21 to α25) tightly packed against the fingers. All of these helices are connected by long loops (Fig. 1a). The α19, η10, η11 region, forms a protrusion that partially blocks the active site cleft (Fig. 1b). Although structurally very different, the location of the C-terminal protrusion in IBDV seems to be reminiscent of the C-terminal initiation platforms of bacteriophage φ6 and hepatitis C virus RDRPs (16, 17).
The Noncanonical Connectivity of the VP1 Palm.
In contrast to the similarities described above, the VP1 palm exhibits an unusual permutation of the sequence motifs A, B, and C from their usual amino- to carboxyl-terminal order A–B–C to the order C–A–B. The structures determined reveal that this rearrangement, previously predicted by computational analyses (2), is compatible with the canonical spatial organization of the major secondary structural elements of the palm fold, maintaining the acidic residues of the polymerase active site in the expected position for catalysis (Figs. 1a and 2 d–f). The permutation of the VP1 palm transforms the βαββαβ fold into a new βββααβ structure (Fig. 2 d–f). In this structure, the central β10–β11 hairpin, containing motif C, and the strand β12, containing motif A, are directly linked and disconnected from helix α13 that contains motif B. These structural alterations result in a different disposition of the region immediately preceding helix α13 (residues 483–490; named B loop from here on) when compared with canonical palms (Fig. 2 d–f). In two of the three structures determined (VP1 apo-form and VP1–Mg2+ complex), the B loop approaches to helix η8 and β11–β12 hairpin, protruding into the active site cleft, and blocking the access of the template and incoming nucleotides. A conformational rearrangement of this region should be required to accommodate these substrates in the catalytic cavity (Fig. 1).
The VP1–Mg2+ Complex.
The incubation of VP1 crystals with GTP and MgCl2 resulted in the incorporation of three Mg2+ ions in the structure. Two of them were found in the active site, bound to the acidic residues Asp-402 (motif C), and Asp-416 and Glu-421 (motif A) in a position close to the expected for the catalytic metal ions (SI Fig. 6). The resulting side-chain organization around the metal ions is very similar to the replication complexes previously reported in canonical polymerases (16, 18, 19). The third metal ion was bound at a site ≈6 Å from the expected catalytic positions, showing a perfect octahedral coordination (SI Fig. 6). A similar finding was reported in the structure of the bacteriophage φ6 (16) and foot-and-mouth disease virus (FMDV) (14) RDRPs. Comparison of IBDV VP1 structures obtained with and without Mg2+ ions did not show any global conformational change in the enzyme on metal binding. No ordered density was observed to position the GTP substrate.
The VP1–VP3 Peptide Complex.
VP1 is enclosed within the capsid, and protein VP3 is the responsible for its encapsidation (9). VP3 also interacts with the viral genome, forming ribonucleoprotein filaments (7). The VP1-binding domain of VP3 has been mapped as the 16 C-terminal-most residues, a region rich in charged amino acids (20).
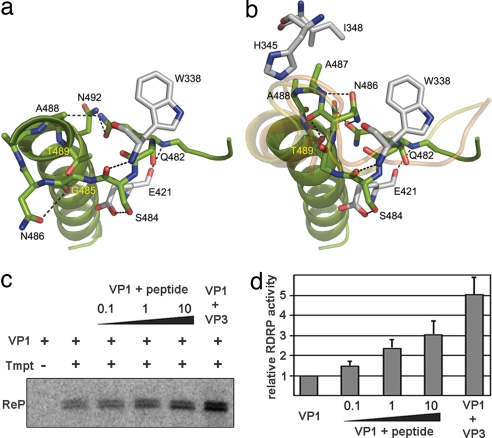
To elucidate the role of VP3 on VP1 binding, we obtained cocrystals of VP1 in complex with the peptide GRLGRWIRTVSDEDLE, representing the VP3 VP1-binding domain. Remarkably, the VP3 peptide binding induced the conformational changes in the B loop required to accommodate the template and incoming nucleotide into the catalytic cavity (Figs. 1 b and c, 3, and 4). In presence of the VP3 peptide, the short helix η8 appears unfolded and the B loop moves ≈8 Å away from its initial position (SI Fig. 7), thus allowing the opening of the active site cavity for substrates entry. The resulting conformation of the B loop after VP3 activation is closely related to that of the equivalent loop in the functional polymerases of FMDV and φ6 (Fig. 3b).
Fig. 3.
Structural rearrangements of the B loop and effects of the VP3 C-terminal peptide and the VP3 polypeptide on VP1 RDRP activity. (a and b) The closed conformation found in the structure of the isolated VP1 (a) and the open conformation found in the VP1–VP3 complex (b). The amino acids involved in intramolecular contacts that stabilize the two alternative conformations of the B loop are shown in sticks and explicitly labeled. The main chain tracing of the equivalent loop in FMDV and φ6 RDRPs correlates well with the open conformation found in the structure of the VP1–VP3 complex (yellow and orange ribbons in b, respectively). (c) Purified hVP1 was preincubated with either the VP3 C-terminal peptide (peptide) or purified hVP3 (VP3). Peptide/hVP1 molar ratios were 0.1:1 (0.1), 1:1 (1), and 10:1 (10), respectively. hVP3 was used at a 2:1 hVP3/hVP1 molar ratio. Samples were supplemented (+) or not (−) with a 544-nt ssRNA+ template (Tmpt). Labeling was performed by using [32P]UTP, and reaction products (ReP) were separated on 5% acrylamide TBE gels. (d) RDRP activity after preincubation with the VP3 C-terminal peptide and the VP3 polypeptide relative to VP1 alone, which was defined as 1. The mean value for each condition was obtained from three independent experiments.
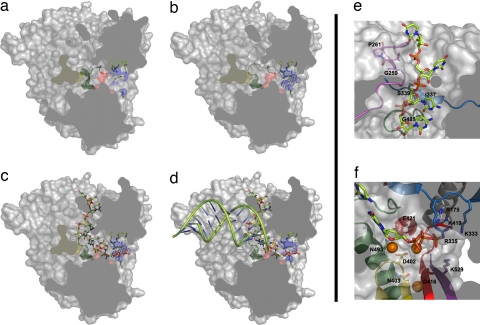
Fig. 4.
A model for activation of the VP1 polymerase and RNA synthesis. A space-filling representation of the enzyme (gray) has been cut to expose the three channels by which the different substrates accede to the active site (red surface). (a) In the absence of VP3, the B loop (green ribbon) partially occludes the binding site of the template base. (b) VP3 accedes to the VP1 active site, through the nucleotide entry tunnel, promoting the conformational change of the B loop that opens the catalytic cavity for template entry. Side chains of basic residues forming the tunnel are shown as sticks and blue surfaces. The electron density corresponding to the visible part of the VP3 peptide is shown as a blue chicken wire. (c) Once the template acceptor base reaches the active site, a second rearrangement should occur, facilitating the exit of VP3 and the entry of rNTP substrates to the active site. (d) After the catalysis of the first phosphodiester bond and pyrophosphate release, the newly synthesized RNA can ratchet down, displacing the C-terminal region (yellow surface). This movement would allow the exit of the nascent duplex out of the polymerase active site. (e) Model of VP1–RNA interactions. The motif G (pink) forms the entry of the template channel. The position of the template acceptor base in the VP1 active site is stabilized by interactions mediated by residues Ile-337 and Ser-339 of motif F (blue) in fingers and Gly-485 of motif B (green) in the palm. (f) The position of the incoming nucleotide appears stabilized by residues Asp-402, Asn-403 (motif C; yellow), Arg-529 (motif D; purple), Lys-333, and Arg-335 (motif F; blue), and residue Arg-175 (helix α5; gray). The docking model also shows how the two Mg2+ ions (orange balls) that coordinate the acidic residues Asp-402 (yellow) and Asp-416 and Asp-421 (red) of the active site are in good position to bind the incoming rNTP.
The VP3 peptide appears poorly ordered in the structure of the complex. Only a weak elongated extra density was detected at the nucleotide entry tunnel (Fig. 4b). This density could be interpreted as due to the presence of ≈5 aa in an extended conformation. The electropositive character of this tunnel, contributed by the basic side chains of Arg-175 (helix α5), Lys-333 and Arg-335 (motif F), Lys-419 (motif A), and Lys-529 (motif D), suggests that the visible density might correspond to the negatively charged C-terminal-most end of VP3 (amino acids DEDLE). Because this tunnel serves also for the entry of incoming ribonucleoside triphosphates (rNTPs) (Fig. 4c) and the exit of the pyrophosphate products, this nucleotide diffusion would probably promote the release of VP3 when the template acceptor base occupies its correct position in the catalytic cavity (Fig. 4d).
The structural observations prompted us to determine the effect of the VP3 C-terminal peptide on the catalytic activity of VP1. For this, VP1 was incubated with different peptide concentrations, and then allowed to replicate an IBDV-derived ssRNA+ template. As shown in Fig. 3 c and d, preincubation of VP1 with the peptide has a significant concentration-dependent effect, reaching a 3-fold increase on VP1 catalytic activity when using 10:1 peptide/VP1 molar ratios. Higher peptide concentrations cause a drastic drop on VP1 activity (data not shown). As expected, preincubation with purified VP3 has even higher boosting effect (up to 5-fold increase) on VP1 polymerase activity (Fig. 3 c and d). These results show that, although VP3 (or the VP3 peptide) is not essential for the initiation of RNA synthesis, it promotes a major enhancement on VP1 catalytic activity in vitro.
Hypothetical Model of RNA Binding and Polymerization.
The structure of VP1 was obtained in absence of RNA. However, the high degree of structural homology observed between VP1, and FMDV and φ6 RDRPs crystallized in presence of different substrates (14, 16) facilitates the superposition of the template-primers and incoming nucleotides on the activated IBDV enzyme. Furthermore, because of the presence of C-terminal protrusions in both VP1 and φ6 RDRPs, we have used the φ6–RNA initiation complex (16) as the best fitting model (Fig. 4c). This model docking allows the identification of the VP1 residues that come into proximity of the ligands, revealing that many strictly conserved amino acids in birnavirus polymerases are in position to interact with the RNA substrates. Most of these putative interacting residues are also conserved in FMDV and φ6 RDRPs (SI Fig. 8). The structural motif G (residues 252–261; within the loop connecting strand β6 and helix η4), highly conserved in the Birnaviridae family (SI Fig. 5), forms the expected template channel entry (Fig. 4e). The equivalent region in picornaviruses contains a 100% conserved cis-Pro (equivalent to Pro-261 in IBDV, also in cis configuration) lining this groove that is preceded by a 100% conserved glycine (equivalent to Gly-259 in IBDV). Mutational experiments in FMDV and poliovirus revealed the critical role of these residues in template binding and nucleic acid translocation during synthesis (21, 22). The φ6 RDRP also has a cis-Pro in an equivalent position but lacks the neighboring Gly. The structural similarities of the region containing motif G in picornavirus, birnavirus, and bacteriophage φ6 suggest similar template binding and RNA translocation modes in the three virus families.
The docking model shows how the two Mg2+ ions bound to the polymerase active site in the VP1–Mg2+ complex (SI Fig. 6) are in good position to bind the incoming rNTP (Fig. 4f). The basic residues of helix α5 and motif F are also in an ideal position to interact with the negatively charged phosphates of the incoming nucleotide (Fig. 4f). In our model, the side chains of Glu-421 (motif A) and Asn-493 (motif B) are in close proximity to both the OH2′ and OH3′ of the rNTP. In picornaviruses, it has been shown that equivalent Asp and Asn residues play a critical role for both rNTP selection and binding (23).
The model also shows how the VP1 C-terminal region, α19–η10–η11, impedes the exit of the nascent dsRNA product. On the formation of the first phosphodiester bond, a new conformational rearrangement would be required to allow the elongation stage of RNA synthesis (Fig. 4 c and d).
Evolutionary Relationships.
The striking structural similarities between FMDV 3D and the IBDV VP1 polymerases suggest a direct evolutionary link between picornavirus and birnavirus RDRPs. The structural similarity of the RDRPs also correlates with other similarities: Protein priming of RNA synthesis with a special viral protein (VPg) was originally discovered in picornaviruses, and all picorna-like viruses may use this mechanism. In birnaviruses, a fraction of the replicase molecules are covalently linked to the 5′ end of genome dsRNAs, and these molecules are likely to be used to prime RNA synthesis (8).
Computational analyses indicate that canonical and permuted palms originated via circular permutation from a single polypeptide ancestor (2). The overwhelming overrepresentation of canonical palms in RDRPs of current virus families, suggests that original polymerases might have possessed a noncanonical palm organization. A single circular permutation event might have led to the assembly of a somewhat more efficient canonical palm configuration that was rapidly fixed and disseminated through the virus world. According to this view, the noncanonical RDRPs of birnaviruses and tetraviruses could be considered as “living fossil” replicases.
The most remarkable biological signature of birnaviruses is the lack of an inner icosahedral (T = 2) transcriptional core, an essential element of particles from any other dsRNA virus family with extracellular phases in their life cycles (5). It has been recently proposed that prototypical dsRNA viruses might have evolved from the merging of a birnavirus precursor (affording the T = 13 capsid) and a totivirus ancestor (providing the T = 2 inner core) (24). Previous data supported the notion that VP3, although incapable of assembling into icosahedral structures, effectively substitutes the T = 2 core roles, providing a scaffold for T = 13 particle assembly (25) and concealing the dsRNA genome from cellular sensors (7). Data presented here suggest that, in addition to the proposed structural role, VP3 plays a functional role interacting with VP1 and regulating its RDRP activity.
Methods
Generation of Recombinant Viruses.
A recombinant baculovirus (rBV) expressing a full-length VP1 polypeptide fused to a 6× histidine tag was generated from the plasmid pBSK/VP1 described in ref. 9. pBSK/VP1 was digested with XhoI, blunt-ended, digested with NotI, and subcloned into the transfer vector pFastBacHTc (Invitrogen) previously digested with EheI and NotI. The resulting plasmid, pFBHTc_VP1, was used to generate an rBV by using the Bac-to-Bac system following the manufacturers' instructions (Invitrogen). A second rBV expressing an his-tagged version of the VP1 polypeptide lacking the 34 C-terminal residues was generated from pFBHTc_VP1. For this, a KpnI fragment from pFBHTc_VP1 was replaced by a PCR-derived DNA fragment containing an artificial stop codon immediately downstream of codon Leu-845 of the VP1 sequence. The PCR was carried out by using pBSK/VP1 as template and the oligonucleotides 5′-CAGGGGCAAGCTGAGACAGC and 5′-GCGCGGTACCTTAGAGAAGAGCGGCCTGGACACC as primers. The DNA was purified, treated with KpnI, and inserted into KpnI-digested pFBHTc_VP1 to produce pFBHTc_VP1Δ846–879, which was used to generate an rBV as described above.
The vaccinia virus recombinant (rVV) VT7LacOI/hVP3 was generated and amplified as described in ref. 26. For this, African green monkey kidney epithelial BSC-1 cells (American Type Culture Collection) were infected with the rVV VT7LacOI (27), and transfected with pVOTE.2/hVP3. pVOTE.2/hVP3 was generated by subcloning a PCR-derived DNA fragment, containing the VP3 coding sequence fused to an N-terminal 6× histidine tag, into the multiple cloning site of the vector pVOTE2 (27). The PCR was performed by using plasmid pFB-Htc-VP3wt (28) as template and the oligonucleotides 5′-GCGCCATATGTCGTACTACCATCACCATCACC and 5′-CCTCTACAAATGTGGTATGGCTG as primers. The resulting DNA fragment was purified, treated with NdeI and SalI, and inserted into pVOTE.2 digested with the same enzymes.
Production and Purification of VP1 and VP3 Polypeptides.
HighFive cells (Invitrogen) were infected with rBVs, harvested at 72 h after infection, washed twice with PBS, resuspended in lysis buffer (50 mM Tris·HCl, pH 8.0, 500 mM NaCl, and 0.1% igepal) supplemented with protease inhibitors (Complete Mini; Roche), and maintained on ice for 20 min. Thereafter, extracts were centrifuged at 13,000 × g for 10 min at 4°C. Supernatants were collected and subjected to metal affinity chromatography (IMAC) purification with a Ni2+ affinity column (HisTrap HP; GE Healthcare). Resin-bound polypeptides were eluted with elution buffer (50 mM Tris·HCl, pH 8.0, 500 mM NaCl, and 250 mM imidazol). hVP1-containing fractions were pooled, dialyzed against lysis buffer lacking igepal, and subjected to a second purification round. hVP1 samples were treated with rTEV (65 μg/mg hVP1; 20 h; 20°C) and dialyzed against a 50 mM Tris, pH 8.0, and 300 mM NaCl buffer. A third Ni2+ affinity column (HisTrap HP; GE Healthcare) was used to purify the untagged VP1 protein. Finally, protein samples were concentrated to a final concentration of ≈10 mg/ml by using Centricon YM-10 filters (Millipore).
The hVP3 polypeptide was expressed in BSC-1 cells infected with VT7LacOI/hVP3 in the presence of the inducer IPTG. At 48 h after infection, cultures were harvested and processed for hVP3 purification following a protocol similar to that used for hVP1.
Crystallization and Data Collection.
Crystals of the recombinant VP1 were obtained by the vapor diffusion method in hanging drops at 20°C, by mixing equal volumes of VP1 (≈10 mg/ml) and the precipitant solution, containing 10–12% PEG3350, 0.3–0.5 M LiNO3, and a pH range between 6.5 and 8.0. Samarium derivative was prepared by soaking native crystals for 30 min in stabilizing solutions containing the crystallization buffer and 3 mM samarium (III) acetate [(CH3CO2)3Sm·3H2O] before freezing. The VP1–GRLGRWIRTVSDEDLE complex was obtained and crystallized as follows: VP1 was mixed with the VP3-derived peptide (1:4 molar ratio) and incubated for 8 h at 4°C before the crystallization trials. Crystals were grown at 20°C from a solution containing 5% PEG3350, 0.4 M LiNO3, and 0.1 M Tris, pH 7.2. VP1–GTP–Mg2+ complexes were obtained by soaking VP1 native crystals in stabilizing solutions containing 2 mM MgCl2 and 2 mM GTP.
Diffraction data were collected from single frozen crystals by using synchrotron radiation at the European Synchrotron Radiation Facility (Grenoble, France) (beamlines BM16, ID23.1, and ID14.2). All diffraction images were processed with programs MOSFLM and SCALA (29).
Structure Determination and Refinement.
The structure was determined by using phase information from multiwavelength anomalous dispersion of a Samarium derivative (peak and remote wavelength data sets) at 3.8-Å resolution. Seven heavy atom sites were determined and refined by using program SHARP (30). Initial phases were applied to a isomorphous data set at 2.8-Å resolution and improved by solvent flattening and histogram matching with DM (29). Experimental maps allowed the automatic identification of several helical elements in the structure by using the helix recognition procedure of ARP/wARP (29). The VP1 initial model (≈400 aa as polyalanine chains) was manually built by using programs O (31) and Coot (32) by using the conserved structural motifs of FMDV and φ6 RDRPs as a guide. This model was then subjected to a preliminary refinement with REFMAC5 (33) and used as a molecular replacement model for the structure determination with AMoRe (34) of the nonisomorphous 2.4-Å native data (SI Table 1). Manual model rebuilding and sequence assignment was alternated with automatic refinement by using REFMAC5. The final model contains 781 residues of 845 in total (Fig. 1). The missing part includes the 26th N-terminal residues of VP1, two flexible regions within the loops connecting the 310 helix η1 and strand β3 (residues 85 and 90) and the α-helices α23–α24 (residues 760 and 765) and the 40 C-terminal residues (from 805 to 845). SDS/PAGE from the VP1 crystals showed that the treatment with rTEV cleaves not only the His-tag but also an additional flexible region of the VP1 protein. The N-terminal sequencing of this species showed an intact amino terminal end, indicating that the cleavage occurred within the 40 C-terminal residues not determined in the crystal structure.
The initial maps for the VP1–GTP–Mg and VP1–GRLGRWIRTVSDEDLE peptide complexes were obtained after a rigid body fitting of the coordinates of isolated VP1 to the new unit cells (SI Table 1). The final refinement cycles resulted in models with free R factors of 26%, 23%, and 25.6% for the isolated, Mg2+, and VP3 peptide complexes, respectively, and good stereochemistry (SI Table 1).
Polymerization Assays.
VP1 polymerization assays were carried out following a protocol described in ref. 6 with minor modifications. Briefly, reaction mixtures containing 1 μg of purified hVP1 and the appropriate amount of the VP3 C-terminal peptide or purified hVP3, were prepared in 20 μl of transcription buffer [100 mM Tris·HCl (pH 7.5), 125 mM NaCl, 4 mM Cl2Mg, 0.01 mM EGTA, 20 units of RNasin, 1 mM ATP, CTP and GTP, and 0.02 mM UTP). Samples were incubated at 20°C for 30 min, supplemented with 5 μl of ssRNA+ template (0.2 mg/ml) diluted in transcription buffer containing 10 μCi of [α-32P]UTP. After incubation at 40°C for 2 h, samples were digested with proteinase K, extracted with phenol/chloroform, ethanol-precipitated, and resuspended in 18 μl of diethyl pyrocarbonate (DEPC)-treated H2O. After adding 3 μl of 6× loading buffer (10 mM Tris·HCl, pH 7.5, 15% Ficoll 400, 50 mM EDTA, 0.4 orange G, 0.03% bromophenol blue, and 0.03% xylene cyanol), samples were heated at 65°C for 5 min, loaded onto 5% acrylamide TBE (90 mM Tris, 64.6 mM boric acid, and 2.5 mM EDTA, pH 8.3) gels. Radioactive signals were detected with a Storm gel imaging system (Molecular Dynamics). Results were analyzed and quantified with ImageQuant software (Molecular Dynamics).
The ssRNA+ template used for the assays was prepared by in vitro transcription, using bacteriophage T7 RNA polymerase (New England Biolabs), of a plasmid harboring a transcriptional cassette, generated by in vitro DNA synthesis (GenScript Corporation), formed by a 554-bp DNA fragment containing the 5′ and 3′ untranslated regions of IBDV segment B flanked by the T7 promoter and the hepatitis δ ribozyme sequences. After transcription, samples were digested with DNase I (Roche), extracted with phenol/chloroform, precipitated with ethanol, and resuspended in DEPC-treated H2O.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Dr. C. Vonrhein for his help in phase determination; Dr. B. Moss (Bethesda, MD) for the VOTE expression system; and Drs. I. Fita, J. L. Carrascosa, and E. Domingo for critical reading of the manuscript. This work was supported by Ministerio de Educacion y Ciencia Grants AGL2003-07189 (to J.F.R.) and BFU2005-02376/BMC (to N.V.). D.G. and J.Q.-A were supported by I3P contracts from Consejo Superior de Investigaciones Cientificas. X-ray data were collected at the European Molecular Biology Laboratory protein crystallography beam lines BM16, ID23.1, and ID14.2 at the European Synchrotron Radiation Facility within a Block Allocation Group (Barcelona). Financial support was provided by the European Synchrotron Radiation Facility.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2PUS, 2QJ1, 2R70, and 2R72).
This article contains supporting information online at www.pnas.org/cgi/content/full/0704447104/DC1.
References
- 1.Koonin EV, Senkevich TG, Dolja VV. Biol Direct. 2006;1:29. doi: 10.1186/1745-6150-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gorbalenya AE, Pringle FM, Zeddam JL, Luke BT, Cameron CE, Kalmakoff J, Hanzlik TN, Gordon KH, Ward VK. J Mol Biol. 2002;324:47–62. doi: 10.1016/S0022-2836(02)01033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van den Berg TP, Eterradossi N, Toquin D, Meulemans G. Rev Sci Tech. 2000;19:509–543. [PubMed] [Google Scholar]
- 4.Delmas B, Kibenge FSB, Leong JC, Mundt E, Vakharia VN, Wu JL. In: Virus Taxonomy. Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball AL, editors. London: Academic; 2004. pp. 561–560. [Google Scholar]
- 5.Bottcher B, Kiselev NA, Stel'Mashchuk VY, Perevozchikova NA, Borisov AV, Crowther RA. J Virol. 1997;71:325–330. doi: 10.1128/jvi.71.1.325-330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Einem UI, Gorbalenya AE, Schirrmeier H, Behrens SE, Letzel T, Mundt E. J Gen Virol. 2004;85:2221–2229. doi: 10.1099/vir.0.19772-0. [DOI] [PubMed] [Google Scholar]
- 7.Hjalmarsson A, Carlemalm E, Everitt E. J Virol. 1999;73:3484–3490. doi: 10.1128/jvi.73.4.3484-3490.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu HT, Si WD, Dobos P. Virology. 2004;322:199–210. doi: 10.1016/j.virol.2004.01.024. [DOI] [PubMed] [Google Scholar]
- 9.Lombardo E, Maraver A, Caston JR, Rivera J, Fernandez-Arias A, Serrano A, Carrascosa JL, Rodriguez JF. J Virol. 1999;73:6973–6983. doi: 10.1128/jvi.73.8.6973-6983.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan J, Vakharia VN, Tao YJ. Proc Natl Acad Sci USA. 2007;104:7385–7390. doi: 10.1073/pnas.0611599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maraver A, Ona A, Abaitua F, Gonzalez D, Clemente R, Ruiz-Diaz JA, Caston JR, Pazos F, Rodriguez JF. J Virol. 2003;77:6438–6449. doi: 10.1128/JVI.77.11.6438-6449.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrer-Orta C, Arias A, Escarmis C, Verdaguer N. Curr Opin Struct Biol. 2006;16:27–34. doi: 10.1016/j.sbi.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Steitz TA. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 14.Ferrer-Orta C, Arias A, Perez-Luque R, Escarmis C, Domingo E, Verdaguer N. J Biol Chem. 2004;279:47212–47221. doi: 10.1074/jbc.M405465200. [DOI] [PubMed] [Google Scholar]
- 15.Ng KK, Pendas-Franco N, Rojo J, Boga JA, Machin A, Alonso JM, Parra F. J Biol Chem. 2004;279:16638–16645. doi: 10.1074/jbc.M400584200. [DOI] [PubMed] [Google Scholar]
- 16.Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI. Nature. 2001;410:235–240. doi: 10.1038/35065653. [DOI] [PubMed] [Google Scholar]
- 17.Leveque VJ, Johnson RB, Parsons S, Ren J, Xie C, Zhang F, Wang QM. J Virol. 2003;77:9020–9028. doi: 10.1128/JVI.77.16.9020-9028.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ago H, Adachi T, Yoshida A, Yamamoto M, Habuka N, Yatsunami K, Miyano M. Structure (London) 1999;7:1417–1426. doi: 10.1016/s0969-2126(00)80031-3. [DOI] [PubMed] [Google Scholar]
- 19.Tao Y, Farsetta DL, Nibert ML, Harrison SC. Cell. 2002;111:733–745. doi: 10.1016/s0092-8674(02)01110-8. [DOI] [PubMed] [Google Scholar]
- 20.Maraver A, Clemente R, Rodriguez JF, Lombardo E. J Virol. 2003;77:2459–2468. doi: 10.1128/JVI.77.4.2459-2468.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson AA, Peersen OB. EMBO J. 2004;23:3462–3471. doi: 10.1038/sj.emboj.7600357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arias A, Agudo R, Ferrer-Orta C, Perez-Luque R, Airaksinen A, Brocchi E, Domingo E, Verdaguer N, Escarmis C. J Mol Biol. 2005;353:1021–1032. doi: 10.1016/j.jmb.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 23.Gohara DW, Crotty S, Arnold JJ, Yoder JD, Andino R, Cameron CE. J Biol Chem. 2000;275:25523–25532. doi: 10.1074/jbc.M002671200. [DOI] [PubMed] [Google Scholar]
- 24.Coulibaly F, Chevalier C, Gutsche I, Pous J, Navaza J, Bressanelli S, Delmas B, Rey FA. Cell. 2005;120:761–772. doi: 10.1016/j.cell.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 25.Saugar I, Luque D, Ona A, Rodriguez JF, Carrascosa JL, Trus BL, Caston JR. Structure (London) 2005;13:1007–1017. doi: 10.1016/j.str.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 26.Earl PL, Moss B. Current Protocols in Molecular Biology. New York: Wiley; 1993. [Google Scholar]
- 27.Ward GA, Stover CK, Moss B, Fuerst TR. Proc Natl Acad Sci USA. 1995;92:6773–6777. doi: 10.1073/pnas.92.15.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kochan G, Gonzalez D, Rodriguez JF. Arch Virol. 2003;148:723–744. doi: 10.1007/s00705-002-0949-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collaborative computational project N. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. [Google Scholar]
- 30.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Acta Crystallogr D Biol Crystallogr. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 31.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 34.Navaza J. Acta Crystallogr D Biol Crystallogr. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.