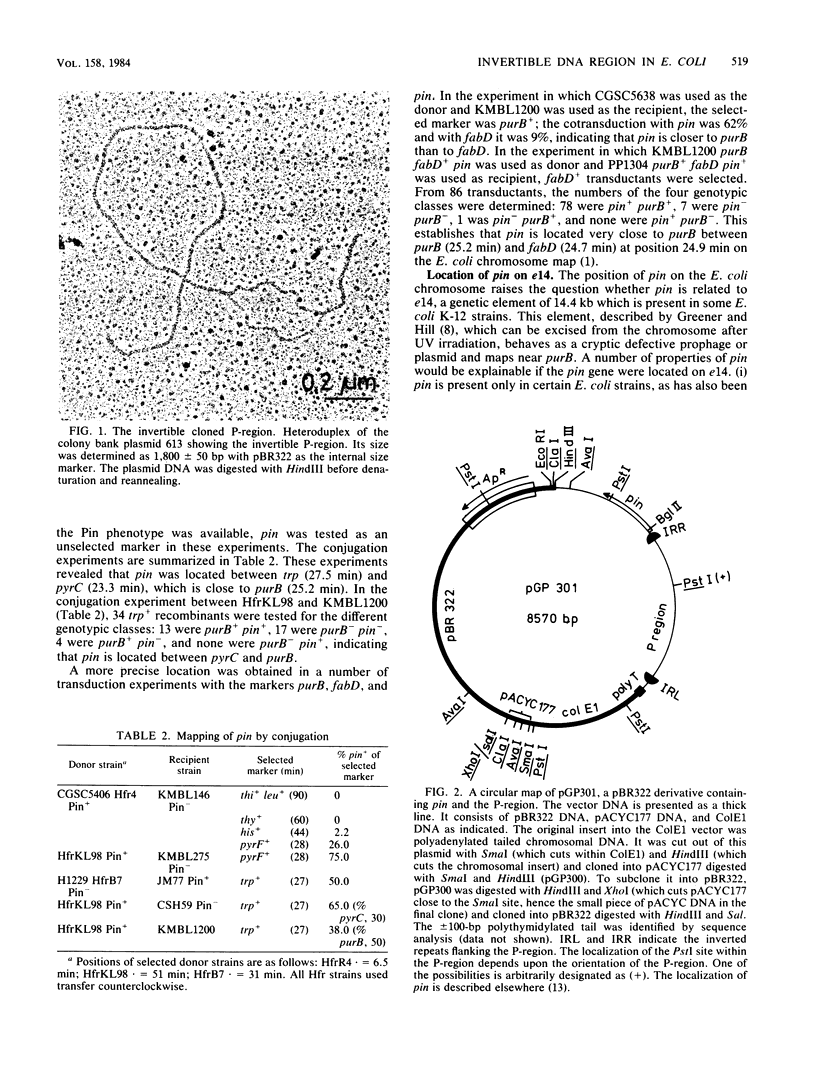
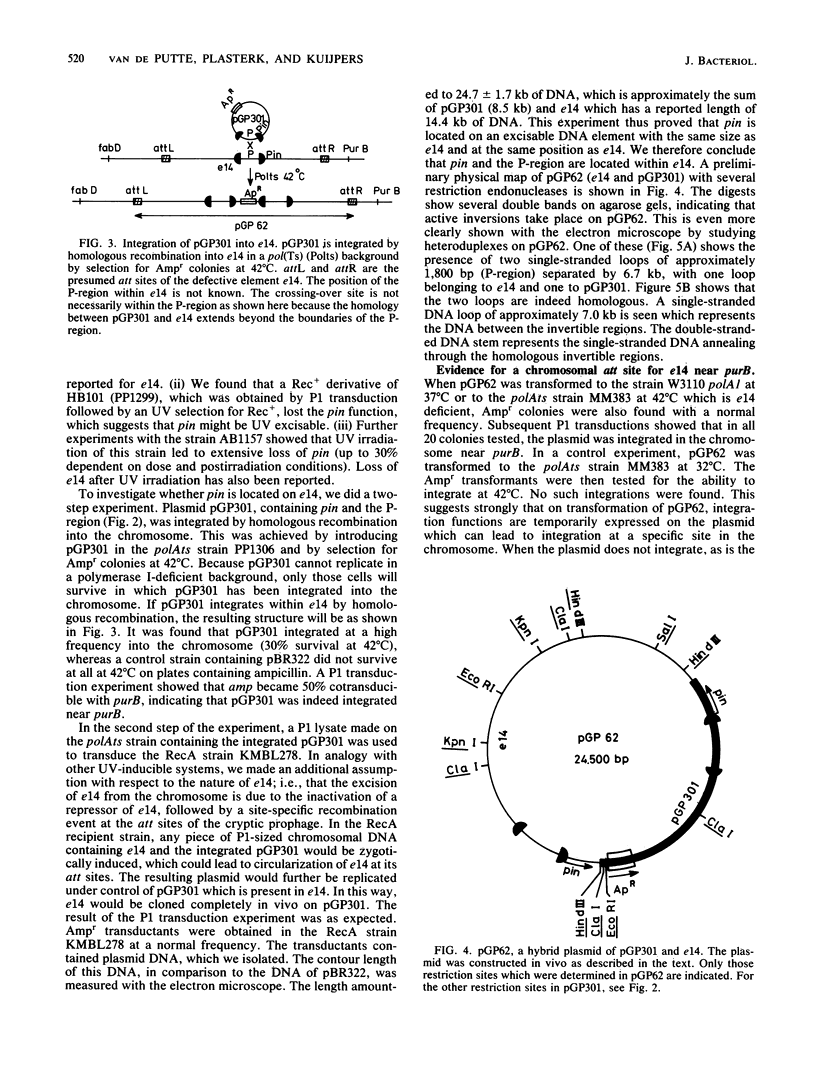
Abstract
The Gin product catalyzes an inversion of 3,000 base pairs of DNA in the genome of bacteriophage Mu. The orientation of the invertible of G-region determines the host range of the phage. Gin- mutants are complemented by a host function in strain HB101 and several other Escherichia coli K-12 strains. At least three clones in the E. coli gene bank described previously (L. Clarke and J. Carbon, Cell 9:91-99, 1976) contained the gin complementing function. This function, which we named pin, catalyzes an inversion of 1,800 base pairs in the adjacent DNA. The invertible region, named the P-region, together with pin, was further subcloned on pBR322. Conjugation and transduction experiments mapped the pin gene between the genes purB and fabD near position 25 on the E. coli chromosome. Also situated in this region is e14, a cryptic, UV- excisable , genetic element (A. Greener and C.W. Hill, J. Bacteriol . 144:312-321, 1980). We demonstrated that pin and the P-region are part of e 14. The e 14 element was cloned on pBR322 by genetic manipulation techniques in vivo. It has the properties of a defective prophage containing integration and excision functions and a SOS-sensitive repressor.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J., Low K. B. Linkage map of Escherichia coli K-12, edition 6. Microbiol Rev. 1980 Mar;44(1):1–56. doi: 10.1128/mr.44.1.1-56.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow L. T., Bukhari A. I. The invertible DNA segments of coliphages Mu and P1 are identical. Virology. 1976 Oct 1;74(1):242–248. doi: 10.1016/0042-6822(76)90148-3. [DOI] [PubMed] [Google Scholar]
- Clarke L., Carbon J. A colony bank containing synthetic Col El hybrid plasmids representative of the entire E. coli genome. Cell. 1976 Sep;9(1):91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- Diderichsen B. flu, a metastable gene controlling surface properties of Escherichia coli. J Bacteriol. 1980 Feb;141(2):858–867. doi: 10.1128/jb.141.2.858-867.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstein B. I. Phase variation of type 1 fimbriae in Escherichia coli is under transcriptional control. Science. 1981 Oct 16;214(4518):337–339. doi: 10.1126/science.6116279. [DOI] [PubMed] [Google Scholar]
- Giphart-Gassler M., Plasterk R. H., van de Putte P. G inversion in bacteriophage Mu: a novel way of gene splicing. Nature. 1982 May 27;297(5864):339–342. doi: 10.1038/297339a0. [DOI] [PubMed] [Google Scholar]
- Greener A., Hill C. W. Identification of a novel genetic element in Escherichia coli K-12. J Bacteriol. 1980 Oct;144(1):312–321. doi: 10.1128/jb.144.1.312-321.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe M. M., O'Day K. J., Schultz D. W. Isolation of mutations defining five new cistrons essential for development of bacteriophage Mu. Virology. 1979 Mar;93(2):303–319. doi: 10.1016/0042-6822(79)90235-6. [DOI] [PubMed] [Google Scholar]
- Kamp D., Kahmann R. The relationship of two invertible segments in bacteriophage Mu and Salmonella typhimurium DNA. Mol Gen Genet. 1981;184(3):564–566. doi: 10.1007/BF00352543. [DOI] [PubMed] [Google Scholar]
- Kamp D., Kahmann R., Zipser D., Broker T. R., Chow L. T. Inversion of the G DNA segment of phage Mu controls phage infectivity. Nature. 1978 Feb 9;271(5645):577–580. doi: 10.1038/271577a0. [DOI] [PubMed] [Google Scholar]
- Kutsukake K., Iino T. Inversions of specific DNA segments in flagellar phase variation of Salmonella and inversion systems of bacteriophages P1 and Mu. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7338–7341. doi: 10.1073/pnas.77.12.7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk R. H., Brinkman A., van de Putte P. DNA inversions in the chromosome of Escherichia coli and in bacteriophage Mu: relationship to other site-specific recombination systems. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5355–5358. doi: 10.1073/pnas.80.17.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk R. H., Ilmer T. A., Van de Putte P. Site-specific recombination by Gin of bacteriophage Mu: inversions and deletions. Virology. 1983 May;127(1):24–36. doi: 10.1016/0042-6822(83)90367-7. [DOI] [PubMed] [Google Scholar]
- Symonds N., Coelho A. Role of the G segment in the growth of phage Mu. Nature. 1978 Feb 9;271(5645):573–574. doi: 10.1038/271573a0. [DOI] [PubMed] [Google Scholar]
- Wijffelman C. A., Westmaas G. C., van de Putte P. Vegetative recombination of bacteriophage Mu-1 in Escherichia coli. Mol Gen Genet. 1972;116(1):40–46. doi: 10.1007/BF00334258. [DOI] [PubMed] [Google Scholar]
- Zieg J., Simon M. Analysis of the nucleotide sequence of an invertible controlling element. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4196–4200. doi: 10.1073/pnas.77.7.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Putte P., Cramer S., Giphart-Gassler M. Invertible DNA determines host specificity of bacteriophage mu. Nature. 1980 Jul 17;286(5770):218–222. doi: 10.1038/286218a0. [DOI] [PubMed] [Google Scholar]