Abstract
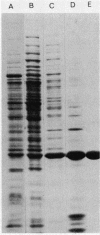
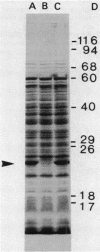
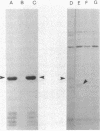
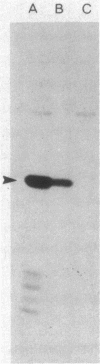
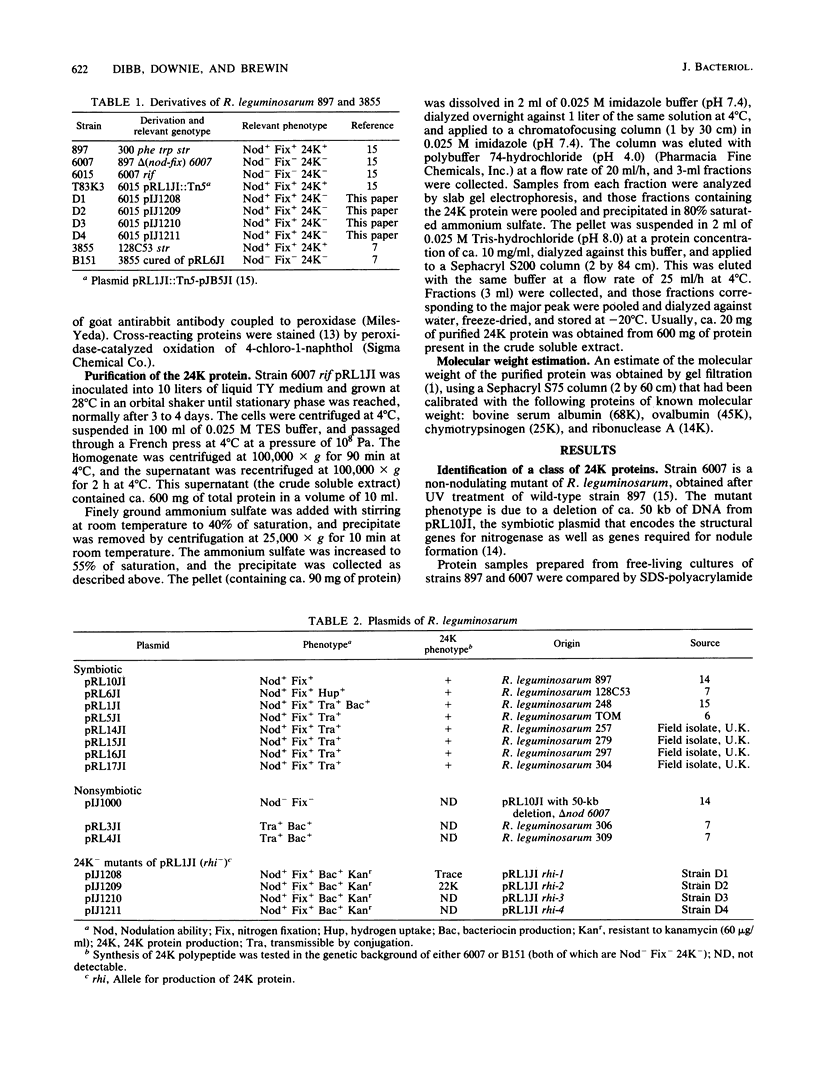
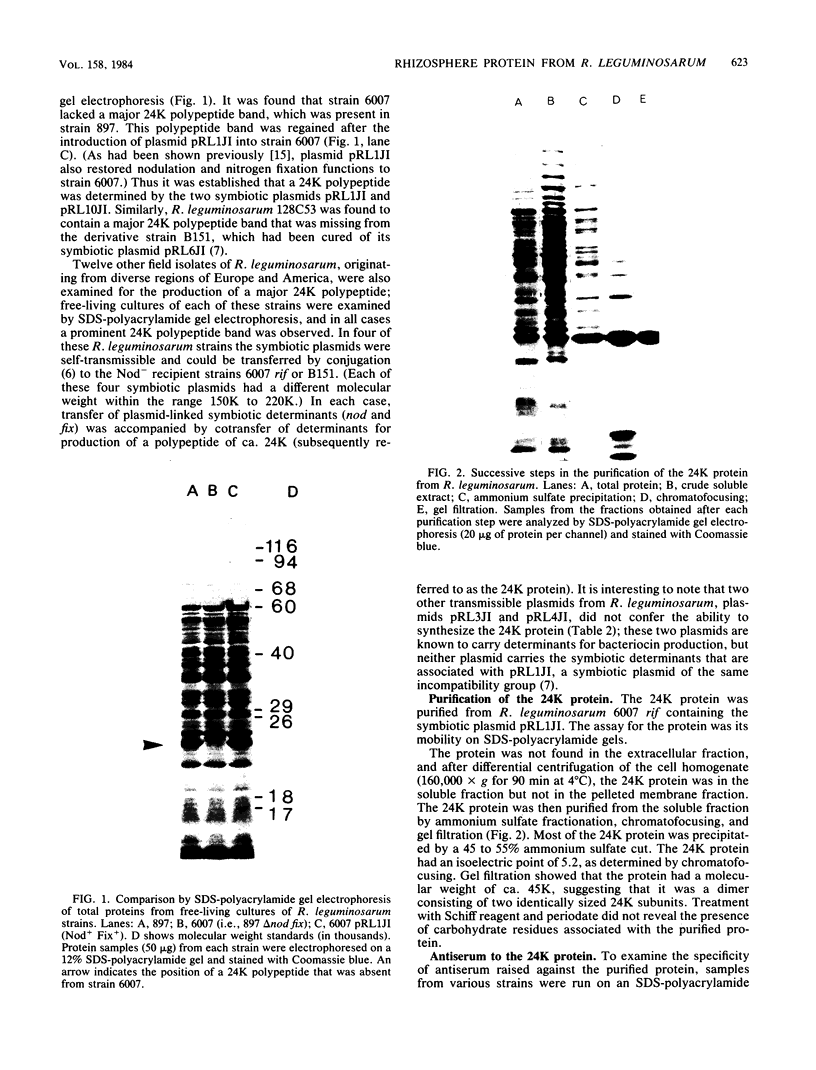
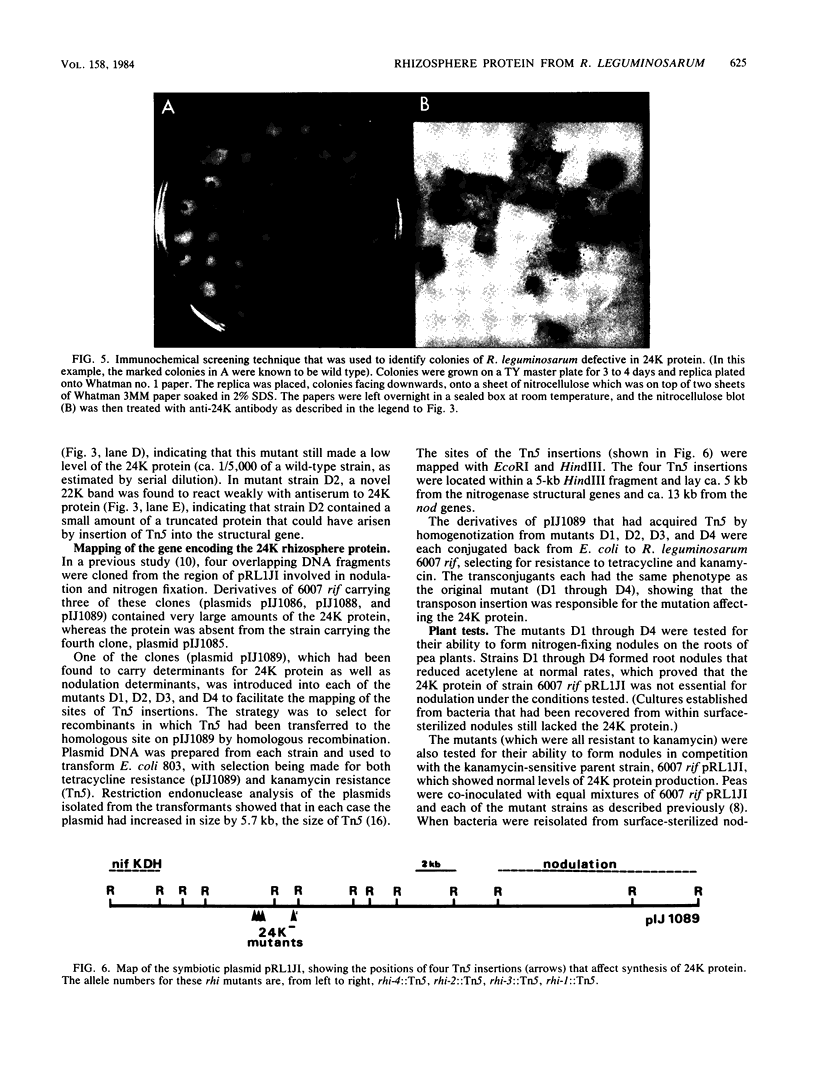
A protein was identified which was made by wild-type strains of Rhizobium leguminosarum but not by nodulation-deficient derivatives which had deletions of their symbiotic plasmids. The protein, which had a subunit molecular weight of ca. 24,000 ( 24K ), was found to be present in large amounts within bacteria that had been reisolated from the surface of inoculated pea roots but was not detected in bacteroids isolated from nodules. The protein could also be induced during growth of R. leguminosarum on nutrient medium and was purified from the cytoplasmic fraction of broken cells. Antiserum raised against the purified protein was used to screen transposon-induced mutants of R. leguminosarum, and four independent mutants were isolated which lacked the protein. The sites of the Tn5 insertions were found to map between the nitrogenase and nodulation genes on symbiotic plasmid pRL1JI , ca. 5 kilobases from the nitrogenase genes and 13 kilobases from the nodulation genes. Genetic determinants for the 24K protein were found to be closely linked to plasmid-borne nodulation genes for all strains of R. leguminosarum tested. However, the mutants which lacked the 24K protein still formed normal nitrogen-fixing nodules on peas, and the function of the protein is unknown.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews P. Estimation of the molecular weights of proteins by Sephadex gel-filtration. Biochem J. 1964 May;91(2):222–233. doi: 10.1042/bj0910222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beringer J. E. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974 Sep;84(1):188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- Bowen B., Steinberg J., Laemmli U. K., Weintraub H. The detection of DNA-binding proteins by protein blotting. Nucleic Acids Res. 1980 Jan 11;8(1):1–20. doi: 10.1093/nar/8.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie J. A., Ma Q. S., Knight C. D., Hombrecher G., Johnston A. W. Cloning of the symbiotic region of Rhizobium leguminosarum: the nodulation genes are between the nitrogenase genes and a nifA-like gene. EMBO J. 1983;2(6):947–952. doi: 10.1002/j.1460-2075.1983.tb01526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt T. A rapid method for the identification of plasmid desoxyribonucleic acid in bacteria. Plasmid. 1978 Sep;1(4):584–588. doi: 10.1016/0147-619x(78)90016-1. [DOI] [PubMed] [Google Scholar]
- Fairbanks G., Steck T. L., Wallach D. F. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry. 1971 Jun 22;10(13):2606–2617. doi: 10.1021/bi00789a030. [DOI] [PubMed] [Google Scholar]
- Hawkes R., Niday E., Gordon J. A dot-immunobinding assay for monoclonal and other antibodies. Anal Biochem. 1982 Jan 1;119(1):142–147. doi: 10.1016/0003-2697(82)90677-7. [DOI] [PubMed] [Google Scholar]
- Jorgensen R. A., Rothstein S. J., Reznikoff W. S. A restriction enzyme cleavage map of Tn5 and location of a region encoding neomycin resistance. Mol Gen Genet. 1979;177(1):65–72. doi: 10.1007/BF00267254. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]