Abstract
The major mutagenic base lesion in DNA caused by exposure to reactive oxygen species is 8-hydroxyguanine (8-oxo-7,8-dihydroguanine). In bacteria and Saccharomyces cerevisiae, this damaged base is excised by a DNA glycosylase with an associated lyase activity for chain cleavage. We have cloned, sequenced, and expressed a human cDNA with partial sequence homology to the relevant yeast gene. The encoded 47-kDa human enzyme releases free 8-hydroxyguanine from oxidized DNA and introduces a chain break in a double-stranded oligonucleotide specifically at an 8-hydroxyguanine residue base paired with cytosine. Expression of the human protein in a DNA repair-deficient E. coli mutM mutY strain partly suppresses its spontaneous mutator phenotype. The gene encoding the human enzyme maps to chromosome 3p25. These results show that human cells have an enzyme that can initiate base excision repair at mutagenic DNA lesions caused by active oxygen.
DNA bases are susceptible to damage in vivo by lipid peroxidation and other endogenous cellular processes that generate reactive oxygen species, as well as by low-level ionizing radiation and oxidative effects of near-ultraviolet (320–380 nm) light. At present there is no direct method to measure the in vivo rate of generation of such endogenous oxidative DNA damage, so its relative importance remains uncertain; recent estimates for mammalian cells have varied from a few hundred to 4 × 105 oxidatively altered DNA base residues being introduced per day in each cell (1, 2). Major expected variations between different cell types and subcellular compartments further complicate the issue. The existence of specific DNA repair enzymes that remove oxidized bases from DNA strongly indicates that this form of endogenous damage is physiologically relevant. DNA pyrimidine residues can be oxidized to noncoding ring-saturated and ring-fragmented derivatives (e.g., thymine glycol, cytosine glycol, and N-substituted urea). A DNA glycosylase with an associated apurinic/apyrimidinic (AP) lyase activity can initiate DNA repair of such lesions, and an Escherichia coli enzyme of this type, endonuclease III, has been extensively characterized (3). A human homolog of E. coli endonuclease III exists, and the cDNA encoding the enzyme has been cloned and functionally expressed (4). In purines, the imidazole ring is more susceptible to oxidative damage than the pyrimidine ring, so DNA purine residues are converted to the ring-saturated and ring-fragmented derivatives, 8-hydroxyguanine and formamidopyrimidines. A distinct DNA glycosylase is present to remove such damage, and the E. coli enzyme excises both types of lesions at similar rates (5). This formamidopyrimidine–DNA glycosylase (Fpg, MutM) was initially discovered by its action on the cytotoxic ring-fragmented purines (6, 7), but the most important substrate for the enzyme appears to be the mutagenic base 8-hydroxyguanine, which usually occurs in its keto form, 8-oxo-7,8-dihydroguanine (8-oxoG) (8).
In spite of the continuous cellular repair of base derivatives generated by endogenous DNA decay, minute amounts of unrepaired DNA damage (up to 1,000 residues per genome), susceptible to treatment with E. coli Fpg or endonuclease III in vitro, have been reported to be present in the DNA of mammalian cells (9). The 8-oxoG residues generate transversion mutations, because they can base pair with adenine residues in an anti-syn arrangement, so active repair serves to reduce spontaneous mutation frequencies (10, 11). A Saccharomyces cerevisiae functional homolog of the E. coli fpg gene, OGG1 (8-oxoguanine DNA glycosylase), has been cloned and characterized (12, 13). In the present work, we describe a human homolog of the microbial genes which encodes an enzyme that removes 8-oxoG residues from DNA. It should now be possible to construct or identify mammalian cells deficient in this form of DNA repair, which will allow improved and quantitative determinations of the rate of endogenous DNA base damage by active oxygen in vivo.
MATERIALS AND METHODS
Bacterial Strains and Reagents.
E. coli strains used for complementation assays were AB1157 and its derivative, MK611 (mutY mutM::Knr) from M. Sekiguchi (Kyushu University, Japan). E. coli Fpg protein was prepared from the overproducer strain JM105/pFPG230 (5). Plasmid pSE420 was from Invitrogen; the expression strain BL21(DE3), plasmid pET28c(+), and T7⋅Tag Antibody HRP conjugate were from Novagen. 8-Oxo-2′-deoxyguanosine-CE phosphoramidite was purchased from Glen Research (Sterling, VA).
Sequence Homology Search and Screening for Full-Length cDNA.
A homology search using the S. cerevisiae OGG1 amino acid sequence against a database containing ≈500,000 human expressed sequence tags (ESTs; Genomic Research and Human Genome Sciences, Rockville, MD) (14, 15) was performed. Candidate ESTs were identified, and approximately 106 recombinants of a human cDNA library were screened with a radiolabeled partial cDNA probe by standard procedures (16). Apparently full-length hOGG1 cDNAs were isolated and subcloned as a SalI–NotI restriction fragment into the multiple cloning site of pSPORT (Life Technologies, Gaithersberg, MD) with the 5′ end proximal to the T7 promoter. The insert size of positive clones was verified by PCR with gene-specific and vector-specific primers. DNA sequencing was carried out on both strands by using an automated ABI 373 DNA Analysis System (Applied Biosystems). The hOGG1 sequence has been deposited in the GenBank database.
Protein Overexpression and Purification.
The 1.3-kb open reading frame coding for the hOGG1 protein was amplified by PCR using Pfu DNA polymerase (Stratagene) and the oligonucleotides 5′-TATGGATCCGAATGCCTGCCCGCGCGCTTC-3′ and 5′-GAGTGCTCGAGACCAGCTCAACAGGAGACT-3′ to engineer BamHI and XhoI restriction sites at the 5′ and 3′ ends of the hOGG1 cDNA, respectively. The amplified fragment was inserted into the pET-28c(+) vector by digestion with BamHI and XhoI restriction enzymes and ligation, resulting in the addition of a 6-histidine tag at the amino-terminal end of the hOGG1 protein. The insert was sequenced on both strands, and products of two independent PCRs were subcloned and sequenced.
The plasmid carrying hOGG1 cDNA was used to transform E. coli BL21(DE3) (17) and transformant colonies inoculated into 3 liters of Luria–Bertani (LB) medium containing kanamycin (30 μg/ml). The culture was incubated with shaking, first at 37°C until A600 reached 0.250 and then at 15°C. After 24 h of incubation at 15°C, the cells were collected by centrifugation at 4,000 g for 30 min at 4°C, and resuspended in 20 ml of buffer (HSB: 20 mM Tris⋅HCl, pH 8.0/0.5 M NaCl/5 mM imidazole/1 mM phenylmethylsulfonyl fluoride). The cells were disrupted by sonication, and the extract was clarified by centrifugation. The supernatant was mixed with 2 ml of Ni2+-nitrilotriacetic acid resin (Qiagen, Chatsworth, CA) that had been preequilibrated with HSB, and stirred gently for 1 h. The resin was then packed into a column and washed with 20 ml of HSB followed by 20 ml of HSB supplemented with 60 mM imidazole. Histidine-tagged protein was eluted with 10 ml of HSB containing 500 mM imidazole and collected in 1-ml fractions. An aliquot of each fraction was analyzed by SDS/PAGE and those containing the overexpressed protein were pooled and dialyzed for 12 h against 50 mM Tris⋅HCl, pH 7.5/30 mM NaCl/1 mM EDTA/1 mM DTT/50% glycerol. For immunoblotting, proteins were separated by SDS/PAGE, transferred to nitrocellulose, and detected with the T7⋅Tag Antibody HRP conjugate by chemiluminescence (ECL, Amersham).
Enzyme Assays.
A 49-mer oligonucleotide containing a single 8-oxoG (5′-TAGACATTGCCATTCTCGATA(8-oxoG)GATCCGGTCAAACCTAGACGAATTCCG-3′) was synthesized on a Perkin–Elmer/Applied Biosystems model 380B DNA synthesizer. The oligomer was deprotected with ammonia solution containing 0.25 M 2-mercaptoethanol and purified by 20% PAGE. The oligonucleotide (3 pmol) was 32P-labeled at the 5′ terminus with T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (Amersham) and annealed to a complementary strand containing either an adenine (A) or a cytosine (C) residue opposite the 8-oxoG residue.
A reaction mixture (15 μl) containing 40 mM Hepes⋅KOH (pH 8.0), 0.1 M KCl, 0.5 mM EDTA, 0.5 mM DTT, 0.2 mg/ml BSA, and 75 fmol of 32P-labeled 8-oxoG-containing duplex was incubated at 37°C for 1 h with purified hOGG1 (1 μg). Reactions were stopped by adding an equal volume of 95% formamide/dyes. The products were separated by gel electrophoresis and visualized by autoradiography or analyzed on a PhosphorImager (Molecular Dynamics). Electrochemical detection of free 8-oxoG in dried ethanol supernatants of 100-μl reaction mixtures (containing phosphate buffer, pH 7, instead of Hepes, and no DTT) was performed with the Beckman HPLC Gold System using an octadecylsilane ABZ plus column (Supelco) and an EG & G (Salem, MA) electrochemical detector model 400.
Assay for Mutation Suppression in E. coli.
The 1.3-kb cDNA encoding hOGG1 was subcloned as a SalI–NotI restriction fragment into the multiple cloning site of the pSE420 vector to place it under the control of an isopropyl β-d-thiogalactopyranoside (IPTG)-inducible promoter. E. coli strains AB1157 (wild type) or MK611 (mutM mutY) were transformed with either the vector or the plasmid containing hOGG1 cDNA. A single transformant was inoculated into 10 ml of LB medium containing carbenicillin (50 μg/ml) and incubated at 37°C for 7 h. The culture was diluted in fresh medium and 150–250 cells were inoculated in 5 ml of LB medium containing carbenicillin in the presence or absence of 1 mM IPTG. Five independent cultures were grown for 24 h at 15°C with shaking and then analyzed for generation of rifampicin-resistant (RifR) mutants after plating on LB agar containing carbenicillin (50 μg/ml) and rifampicin (100 μg/ml).
Chromosomal Mapping.
The hOGG1 cDNA was labeled with digoxigenin-dUTP by nick-translation (Boehringer Mannheim), and fluorescence in situ hybridization was performed according to Johnson et al. (18). Individual chromosomes were counterstained with DAPI (4′,6-diamidino-2-phenylindole), and color digital images, containing both DAPI and gene signals, were recorded using a triple-band pass filter set (Chroma Technology, Brattleboro, VT) in combination with a charged coupled-device camera (Photometrics, Tucson, AZ) and variable excitation wavelength filters (19). Images were analyzed by using the isee software package (Inovision, Durham, NC).
RESULTS
Human cDNA Encoding 8-oxoG-DNA Glycosylase.
A database search of amino acid sequences conserved between several bacterial formamidopyrimidine-DNA glycosylases did not reveal a similar human sequence. However, when the amino acid sequence of S. cerevisiae OGG1 was used, an EST derived from human frontal cortex was identified that showed partial homology with a region of the yeast OGG1. Other positive ESTs were identified from human keratinocytes, liver, prostate, fetal heart, and the Burkitt lymphoma Raji cell line. Several human cDNA libraries were screened by hybridization with the first EST clone to obtain a full-length human OGG1 (hOGG1) cDNA. Two identical, apparently full-length cDNA sequences were isolated from libraries of a teratocarcinoma cell line and frontal cortex (mainly dendritic cells). The nucleotide sequence of the hOGG1 cDNA revealed a 1,275-bp open reading frame encoding a predicted 47.2-kDa protein of 424 amino acids. The amino acid sequence of hOGG1 showed 33% identity and 54% similarity to S. cerevisiae OGG1, and several highly conserved regions were present (Fig. 1). A notable feature of the hOGG1 sequence is the presence of a putative DNA-binding helix–hairpin–helix motif, previously identified in several different DNA glycosylases (3, 13, 21). Northern blot analysis of several tissues gave very weak signals, and direct enzyme assays of cell extracts also indicated that hOGG1 is expressed at a low level in many cell types (data not shown).
Figure 1.
Sequence homology between human 8-oxoG-DNA glycosylase and S. cerevisiae OGG1. Amino acid sequences were aligned by the pileup and bestfit programs in the Genetics Computer Group package (Version 7; ref. 20). Amino acids identical in both sequences are boxed. The position of the possible helix–hairpin–helix motif is indicated. The Lys and Asp residues that have been associated with dual DNA glycosylase/AP lyase activity (13) are shown with an arrow and a circle, respectively.
Overexpression and Purification of hOGG1.
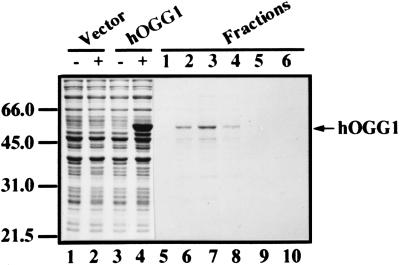
A polyhistidine (His6) affinity tag was attached to the amino terminus of hOGG1 by subcloning into the pET-28c vector. The protein was expressed at a high level following IPTG induction of E. coli BL21(DE3) transformed with the plasmid that carries the hOGG1 cDNA (Fig. 2, lanes 1–4). When the induction was performed at 37°C, 30°C, or 20°C, the protein was insoluble and found in inclusion bodies. However, growing the cells at 15°C for 24 h in the absence of IPTG resulted in low-level expression, with part of the protein remaining soluble. The hexahistidine-tagged hOGG1 protein was purified by Ni2+ affinity chromatography. Elution with 500 mM imidazole revealed a major protein band by SDS/PAGE migrating with an apparent molecular mass of approximately 50 kDa (Fig. 2, lanes 5–10).
Figure 2.
Purification of recombinant hOGG1 protein. The hOGG1 was overexpressed in E. coli, and proteins were visualized on a 10% SDS/polyacrylamide gel by Coomassie blue staining. Lanes 1–4 show overexpression of the protein at 37°C by IPTG induction and contain whole-cell lysate (5 μl) from bacteria harboring the vector before (lane 1) and after induction (lane 2), or the plasmid carrying hOGG1 cDNA before (lane 3) and after induction (lane 4). Lanes 5–10 show peak fractions (10 μl) eluted with 0.5 M imidazole from a Ni2+-nitrilotriacetic acid column loaded with a crude extract from cells grown at 15°C (see Materials and Methods). The few protein bands migrating below hOGG1 represent degradation or premature termination products of the overexpressed protein, as determined by immunoblotting with the T7⋅Tag Antibody (data not shown). Positions of protein size markers in kDa (Bio-Rad) are indicated on the left.
Substrate Specificity of the hOGG1 Protein.
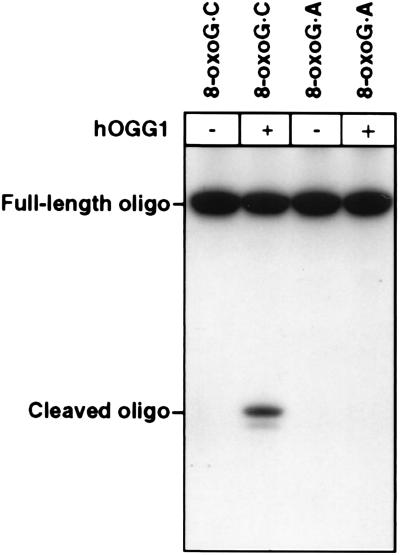
E. coli Fpg protein and S. cerevisiae OGG1 act as DNA glycosylases/AP lyases at 8-oxoG residues base-paired with C, but only very poorly when base-paired with A (10, 12, 13). An 8-oxoG⋅A base pair is instead a substrate for the distinct MutY DNA glycosylase, which excises the adenine residue (10). The hOGG1 protein was found to cleave an 8-oxoG-containing oligonucleotide with a complementary strand that had a C opposite the oxidized residue, although activity was low (Fig. 3). The observed activity was shown to be directly proportional to enzyme concentration over a 10-fold range (data not shown). When the complementary strand instead had an A opposite the 8-oxoG residue, no enzyme activity was detected (Fig. 3). In parallel experiments with E. coli Fpg protein, there was a 100-fold difference in the rate of enzyme attack on these two substrates (data not shown); a similar reduced activity on the substrate with an 8-oxoG⋅A pair by hOGG1 would not have been detected here. When hOGG1 activity on the substrate with an 8-oxoG⋅C base pair was assessed by HPLC with electrochemical detection of 8-oxoG released in free form, the enzyme was 5–10 times more active, indicating incomplete AP lyase activity to generate chain breaks at abasic sites (data not shown). 8-oxo-G was also released in free form by hOGG1 from plasmid DNA treated with methylene blue and light to introduce 8-oxoG residues. The enzyme was at least 10-fold less active on a DNA substrate with formamidopyrimidine instead of 8-oxoG, whereas E. coli Fpg protein was similarly active with both substrates (data not shown). The S. cerevisiae OGG1 was observed previously to act 12 times slower at formamidopyrimidines than at 8-oxoG residues (12). Data on the S. cerevisiae homolog of endonuclease III indicate that this enzyme, rather than OGG1, may be responsible for excision of oxidatively fragmented purine residues from yeast DNA (22). Our hOGG1 preparation was not detectably active on a double-stranded polynucleotide containing pyrimidines fragmented by KMnO4 treatment (4), which is the preferred substrate for the human endonuclease III (data not shown).
Figure 3.
Comparison of hOGG1 activity on double-stranded oligonucleotides containing an 8-oxoG⋅C or an 8-oxoG⋅A mismatch. Oligonucleotides were 32P-labeled at the 5′ end of the 8oxoG-containing strand and were incubated at 37°C for 1 h with purified hOGG1 as described in Materials and Methods. Reaction products were analyzed by autoradiography after electrophoretic separation in a denaturing 20% polyacrylamide gel.
The hOGG1 activity differed markedly from that of E. coli Fpg with regard to salt dependence; when KCl was excluded from the standard reaction mixture with an 8-oxoG-containing substrate, hOGG1 was slightly (1.3-fold) stimulated, whereas E. coli Fpg activity was reduced 3-fold.
Suppression of the Mutator Phenotype of E. coli mutM mutY by hOGG1.
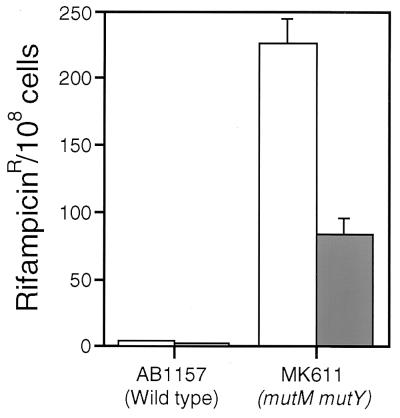
An E. coli mutM(fpg) mutY double mutant showed a high level of spontaneous transversion mutations (10). The hOGG1 cDNA was expressed in E. coli MK611 (mutM mutY) and tested for its ability to suppress the mutator phenotype. The hOGG1 cDNA had been subcloned under control of a tightly repressed promoter that was inducible by IPTG. MK611 cells transformed with hOGG1 showed marked but incomplete suppression of the mutator phenotype (Fig. 4). This decrease was not observed in the absence of IPTG, so the antimutator effect was correlated with expression of the human protein. Similar partial complementation of E. coli mutM mutY with S. cerevisiae OGG1 has been observed previously (12).
Figure 4.
Antimutator effect of hOGG1 cDNA in E. coli (mutM mutY). AB1157 (wild type) or MK611 (mutM mutY) cells transformed with pSE420 (open bars) or pSE420 carrying the hOGG1 cDNA (solid bars) were grown in the presence of 1 mM IPTG and plated both in the presence and absence of rifampicin. Results are from five independent experiments, and error bars are shown.
Chromosomal Localization of hOGG1.
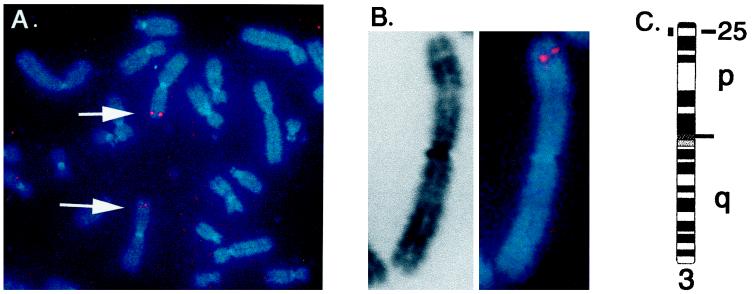
To determine the chromosomal location of the hOGG1 gene, single-gene fluorescence in situ hybridization to human chromosome metaphase spreads was performed (23). Approximately seven spreads were analyzed by eye, most of which had a doublet signal characteristic of genuine hybridization on at least one chromosome 3. Doublet signals were not consistently detected on any other chromosome. Detailed analysis of 12 individual chromosomes, using fluorescence banding combined with high-resolution image analysis, indicated that the hOGG1 gene is positioned within band 3p25. The majority of signals seen were positioned within the distal portion of this band, indicating that the gene may fall within subbands 3p25.2–25.3 (Fig. 5). The DNA repair gene XPC (xeroderma pigmentosum, complementation group C) also is located in this band (24) but is distinct from hOGG1.
Figure 5.
Fluorescence in situ hybridization mapping of the hOGG1 gene. The hOGG1 cDNA was hybridized to normal human male chromosomes. (A) A chromosome spread from a single cell, showing hybridization to the p arm of each chromosome 3 (arrows). (B) Example of a single chromosome 3 with hybridization signal. Background signals in nonchromosomal areas were suppressed in the electronic image for both A and B. (C) Idiogram of chromosome 3, showing the position of the hOGG1 gene.
DISCUSSION
DNA damage caused by exposure to reactive oxygen species such as hydroxyl radical (OH⨪), singlet oxygen (1O2), superoxide radical (O2⨪), and hydrogen peroxide (H2O2) is believed to be one of the most important reasons for endogenous DNA decay. The widespread occurrence of relevant repair enzymes (i.e., DNA glycosylases specific for oxidized bases) reinforces this view. Here we reported on the cloning of a human cDNA encoding an enzyme that acts specifically at mutagenic 8-oxoG residues in DNA. This DNA glycosylase shares some important features with the well characterized E. coli Fpg protein but is more closely related to the recently described S. cerevisiae OGG1 protein (12, 13), so it is called hOGG1. A human cDNA encoding hOGG1, also located to chromosome 3p25, has been independently isolated by J. P. Radicella and S. Boiteux (personal communication). We have shown that the specificity of the hOGG1 enzyme is similar to that of the bacterial and yeast enzymes, cleaving a double-stranded oligonucleotide at an 8-oxoG residue base paired with a C residue, but not detectably at an 8-oxoG base paired with an A (10, 12, 13). By analogy with the known reaction mechanisms of several DNA glycosylases (3–8, 13), the action of hOGG1 apparently involves hydrolytic excision of the free oxidized base, followed by inefficient DNA chain cleavage, presumably by a β-elimination reaction. Moreover, expression of the protein in E. coli partially suppresses the spontaneous mutator phenotype of a mutM(fpg) mutY mutant, which is deficient in repair of endogenously produced 8-oxoG.
Ames et al. (25, 26) have proposed that oxidative DNA damage may be a significant causative factor for cancer and aging in mammals. To evaluate the possible degenerative effects of endogenously produced active oxygen, it would be desirable to generate knockout mice deficient in DNA repair enzymes that counteract oxidative damage. Cloning of mammalian cDNAs encoding such enzymes is a necessary first step in this direction, and the antimutator factor characterized here should be of major relevance in this regard. Previous attempts to generate knockout mice that are deficient in the later steps of base excision repair have resulted in embryonic lethal phenotypes (reviewed in ref. 27), but the properties of microbial mutants deficient in individual DNA glycosylases suggest that loss of such a function may not be a lethal event in mammals.
The hOGG1 gene maps to the short arm of chromosome 3, within the distal portion of band p25 (Fig. 5). Chromosomal abnormalities in this region, including allele loss and deletions, have been reported for several carcinomas, including lung cancer (28). A search for the possible absence of a functional hOGG1 gene in tumors and tumor cell lines with 3p25 alterations should now be undertaken. Such cells might be expected to exhibit an increased spontaneous mutation frequency and genomic instability, which could contribute to malignancy.
Acknowledgments
We thank Iain Goldsmith and the oligonucleotide synthesis laboratory at the Imperial Cancer Research Fund for the synthesis of oligonucleotide substrates, and Graham Daly for purified E. coli Fpg protein. The nucleotide sequence of hOGG1 was determined at the sequencing core facility at Human Genome Sciences Inc. The human tissues used for cDNA library construction were provided by the Cooperative Human Tissue Network, funded by the National Cancer Institute. T.R.A. and A.K. were supported by European Community training fellowships under the Human Capital and Mobility Programme, and C.A. had a long-term European Molecular Biology Organization fellowship.
ABBREVIATIONS
- AP
apurinic/apyrimidinic
- EST
human expressed sequence tag
- IPTG
isopropyl β-d-thiogalactopyranoside
- LB medium
Luria–Bertani medium
- 8-oxoG
8-oxo-7,8-dihydroguanine
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. Y13277).
References
- 1.Lindahl T. Nature (London) 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Blount B C, Mack M M, Wehr C M, MacGregor J T, Hiatt R A, Wang G, Wickramasinghe S N, Everson R B, Ames B N. Proc Natl Acad Sci USA. 1997;94:3290–3295. doi: 10.1073/pnas.94.7.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thayer M M, Ahern H, Xing D, Cunningham R P, Tainer J A. EMBO J. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aspinwall R, Rothwell D G, Roldán-Arjona T, Anselmino C, Ward C J, Cheadle J P, Sampson J R, Lindahl T, Harris P C, Hickson I D. Proc Natl Acad Sci USA. 1997;94:109–114. doi: 10.1073/pnas.94.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karakaya A, Jaruga P, Bohr V A, Grollman A P, Dizdaroglu M. Nucleic Acids Res. 1997;25:474–479. doi: 10.1093/nar/25.3.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chetsanga C J, Lindahl T. Nucleic Acids Res. 1979;6:3673–3684. doi: 10.1093/nar/6.11.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boiteux S, O’Connor T R, Lederer F, Gouyette A, Laval J. J Biol Chem. 1990;265:3916–3922. [PubMed] [Google Scholar]
- 8.Tchou J, Kasai H, Shibutani S, Chung M H, Laval J, Grollman A P, Nishimura S. Proc Natl Acad Sci USA. 1991;88:4690–4694. doi: 10.1073/pnas.88.11.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins A R, Dusinská M, Gedik C M, Stetina R. Environ Health Perspect. 1996;104:465–469. doi: 10.1289/ehp.96104s3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michaels M L, Miller J H. J Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oda Y, Uesugi S, Ikehara M, Nishimura S, Kawase Y, Ishikawa H, Inoue H, Ohtsuka E. Nucleic Acids Res. 1991;19:1407–1412. doi: 10.1093/nar/19.7.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Kemp P A, Thomas D, Barbey R, de Oliveira R, Boiteux S. Proc Natl Acad Sci USA. 1996;93:5197–5202. doi: 10.1073/pnas.93.11.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nash H M, Bruner S D, Schärer O D, Kawate T, Addona T A, Spooner E, Lane W S, Verdine G L. Curr Biol. 1996;6:968–980. doi: 10.1016/s0960-9822(02)00641-3. [DOI] [PubMed] [Google Scholar]
- 14.Adams M D, Kelley J M, Gocayne J D, Dubnick M, Polymeropoulos M H, Xiao H, Merril C R, Wu A, Olde B, Moreno R F. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- 15.Adams M D, Dubnick M, Kerlavage A R, Moreno R F, Kelley J M, Utterback T R, Nagle J W, Fields C, Venter C. Nature (London) 1992;355:632–634. doi: 10.1038/355632a0. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 17.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 18.Johnson C V, Singer R H, Lawrence J B. Methods Cell Biol. 1991;35:73–99. [PubMed] [Google Scholar]
- 19.Johnson C V, McNeil J A, Carter K C, Lawrence J B. Gene Anal Tech Appl. 1991;8:75–76. doi: 10.1016/1050-3862(91)90052-s. [DOI] [PubMed] [Google Scholar]
- 20.Genetics Computer Group. Program manual of the gcg package. Madison, WI: Genetics Computer Group; 1991. , Version 7. [Google Scholar]
- 21.Roldán-Arjona T, Anselmino C, Lindahl T. Nucleic Acids Res. 1996;24:3307–3312. doi: 10.1093/nar/24.17.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eide L, Bjøras M, Pirovano M, Alseth I, Berdal K G, Seeberg E. Proc Natl Acad Sci USA. 1996;93:10735–10740. doi: 10.1073/pnas.93.20.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrence J B, Villnave C A, Singer R A. Cell. 1988;52:51–61. doi: 10.1016/0092-8674(88)90530-2. [DOI] [PubMed] [Google Scholar]
- 24.Wood R D. Annu Rev Biochem. 1996;65:135–167. doi: 10.1146/annurev.bi.65.070196.001031. [DOI] [PubMed] [Google Scholar]
- 25.Ames B N, Shigenaga M K, Hagen T M. Proc Natl Acad Sci USA. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ames B N, Gold L S, Willet W C. Proc Natl Acad Sci USA. 1995;92:5255–5258. doi: 10.1073/pnas.92.12.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kubota K, Nash R A, Klungland A, Schär P, Barnes D E, Lindahl T. EMBO J. 1996;15:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 28.Naylor S, Carritt B. Cytogenet Cell Genet. 1991;58:170–230. [Google Scholar]