Abstract
A gene mutated in the human genetic disorder ataxia-telangiectasia (A-T), ATM, was recently identified by positional cloning. ATM is a member of the phosphatidylinositol-3-kinase superfamily, some of which are protein kinases and appear to have important roles in cell cycle control and radiation signal transduction. We describe herein, to our knowledge, for the first time, the cloning of a full-length cDNA for ATM and correction of multiple aspects of the radio-sensitive phenotype of A-T cells by transfection with this cDNA. Overexpression of ATM cDNA in A-T cells enhanced the survival of these cells in response to radiation exposure, decreased radiation-induced chromosome aberrations, reduced radio-resistant DNA synthesis, and partially corrected defective cell cycle checkpoints and induction of stress-activated protein kinase. This correction of the defects in A-T cells provides further evidence of the multiplicity of effector functions of the ATM protein and suggests possible approaches to gene therapy.
The human genetic disorder ataxia-telangiectasia (A-T) is characterized by neurological degeneration, immunodeficiency, and cancer predisposition (1). Cellular features include chromosomal instability (2), hypersensitivity to ionizing radiation (3, 4), aberrant regulation of DNA synthesis after irradiation (5, 6), and defective cell cycle checkpoints (7, 8). Anomalies in cell cycle control in A-T cells after irradiation were first revealed in the form of radio-resistant DNA synthesis (5, 6). This was followed by the observation that irradiated A-T cells were less delayed in their entry into mitosis than control cells (8). This failure of radiation exposure to delay the entry of A-T cells into mitosis was also extended to the G1/S-phase checkpoint, and in the longer term, A-T cells irradiated in either G1 or S phases were blocked irreversibly at G2/M phase (7). Kastan et al. (9) provided an explanation for the defective G1/S checkpoint when they showed that the radiation induction of p53 was defective in A-T cells, this was subsequently confirmed in several other reports (10–13). It is now evident that the radiation signal transduction pathway operating through p53, WAF1, and the cyclin E–cdk2 complex is defective in A-T cells (14, 15). Furthermore, it has recently been shown that several cyclin-dependent kinases are resistant to radiation inhibition in A-T cells and this appears to be due to insufficient induction and binding of WAF1, suggesting that the defect in the p53 pathway is at least in part responsible for the defect at several cell cycle checkpoints (16).
Functional complementation of the radio-sensitive phenotype in A-T with genomic and cDNA has led to the identification of several genes that correct at least some aspects of this phenotype (17–20), but none of these genes were shown to map to the locus of the A-T gene on chromosome 11 (21). The gene mutated in A-T, ATM, was eventually identified by positional cloning (22). This gene occupies 150 kb of genomic DNA and encodes an mRNA of 13 kb, and the predicted product from the 9.168-kb open reading frame (ORF) is a 350-kDa protein (23). The ATM protein is related to a family of proteins through a C-terminal phosphatidylinositol-3-kinase domain (24–26). Members of the phosphatidylinositol-3-kinase family of proteins are involved in cell cycle control, DNA replication, recombination, and repair (27–31). Savitsky et al. (22) reported the isolation of a partial ATM cDNA (5.9 kb), and the cloning of a DNA contig spanning the complete ORF of the ATM gene was subsequently described (32). We report herein the construction of the first full-length ATM cDNA clone and the use of this clone to correct multiple aspects of the cellular phenotype of A-T.
MATERIALS AND METHODS
Cloning of the Full-Length ATM cDNA in Epstein–Barr Virus (EBV)-Based Shuttle Vectors.
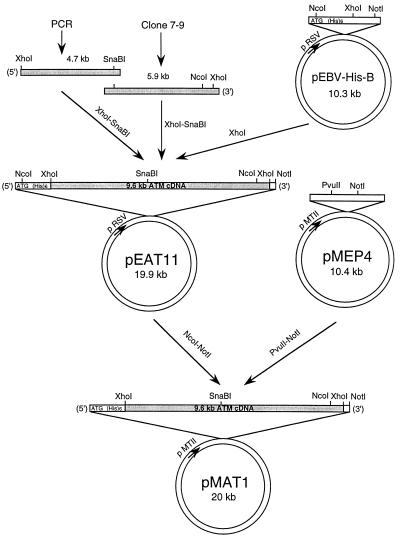
ATM λ clone 7–9 (22), which contains a partial 5.9-kb ATM cDNA fragment, was converted to a plasmid construct (pATM7–9) by a standard process (λ Zap automatic excision process, CLONTECH). mRNA from normal lymphoblastoid cells was extracted, reverse-transcribed into cDNA, and used for PCR amplification of a fragment overlapping the 5′ end of the ATM sequence. Two primers were designed. The first 5′-AGCTCGAGATCACTTAATGATCTGCTTATC-3′ incorporating an XhoI site overlaps the unique ScaI site of the ATM sequence. The reverse primer (5′-CAACATGGTCAGGAAAAGGATC-3′) was located 4686 bp downstream of the ATG start codon of the ATM gene. For PCR (97°C for 3 min, 56°C for 2 min, and 70°C for 4 min, first cycle; 95°C for 30 sec, 56°C for 1 min, and 70°C for 4 min, for cycles 2–32), Expand high-fidelity PCR system (Boehringer Mannheim) was used to minimize the risk of misincorporation. A 4.7-kb ATM fragment was amplified. Both plasmid pATM7–9 and the PCR-amplified 4.7-kb ATM fragment were double-digested with XhoI and SnaBI and fragments of 4.46 kb and 5.12 kb were gel-purified from the digestions, respectively. These two fragments were ligated to each other and the resulting 9.58-kb full-length fragment was cloned into the XhoI site of an EBV-based plasmid vector pEBV-His-B (Invitrogen). The ATM construct was designated pEAT11. In this construct, a 6×His tag sequence was located upstream of the ATM gene. Both the 6×His tag and the ATM gene were subsequently subcloned into another EBV-based inducible vector pMEP4 (33). Briefly, plasmid pEAT11 was partially digested with NcoI (to avoid digestion of the NcoI site in the ATM gene) and blunt-ended with T4 DNA polymerase. A second digestion with NotI was carried out and the 9.58-kb NcoI–NotI ATM fragment was isolated and directionally cloned into plasmid pMEP4, which was double-digested with PvuII and NotI. The generated ATM plasmid construct was designated pMAT1. In both cases DNA sequence analysis was carried out by using a dye terminator kit (Applied Biosystems) to verify the integrity of the constructs.
Cell Culture.
AT1ABR, AT3ABR, and L3 are EBV-transformed A-T lymphoblastoid cell lines, and C3ABR was used as a control lymphoblastoid line. Cells were grown in RPMI 1640 medium supplemented with 10% fetal calf serum under an atmosphere of 5% CO2/95% air. Transfected cells were cultured in the same medium containing hygromycin at 0.2 mg/ml.
Transfection of Human Lymphoblast Cells.
Transfection of EBV-transformed lymphoblastoid cells was carried out as described (20, 34). Briefly, 2 × 106 exponentially growing cells were pelleted, rinsed twice with fetal calf serum-free medium (Opti-MEM, GIBCO/BRL), and resuspended in 1 ml of Opt-MEM medium containing 6 μg of plasmid DNA and 20 μg of Lipofectin (GIBCO/BRL). After incubation for 6–7 h at 37°C, 4 ml of serum-containing RPMI 1640 medium was added and the cells were further incubated. Selection for resistant cells was begun 48 h after transfection with hygromycin B (Boehringer Mannheim) at 0.2 mg/ml. Stably transfected cells are usually obtained at 3–4 weeks after transfection.
Cell Survival.
ATM cDNA expression vector pMAT1 transfected cells were induced or mock-induced for ATM expression with 5 μM CdCl2 for 16 h. The cells were pelleted, suspended in RPMI 1640 medium, and plated out at 2 × 105/ml in culture plates and irradiated with 4 Gy (2.8 Gy/min) of γ-rays. Cell viability was determined by adding 0.1 ml of 0.4% trypan blue to a 0.5-ml cell suspension (20). The number of viable cells were counted up to 4 days after irradiation. Nontransfected cells were used as controls.
Induced Chromosome Aberrations.
Cells were irradiated with 1 Gy of γ-rays. For G2-phase cells, Colcemid (final concentration, 0.1 mg/ml) was added immediately after irradiation, 1–2 h prior to harvesting. The cells were treated for 15 min in 0.075 M KCl, fixed in methanol/glacial acetic acid, 3:1 (vol/vol), and spread on glass slides. The cells were then stained with Giemsa and 50 metaphases were analyzed for each sample (20).
Western Blotting and Immunoprecipitation.
Cells were harvested and lysed in RIPA lysis buffer (50 mM Tris⋅HCl, pH 7.6/150 mM NaCl/1% Triton X-100/0.1% SDS/0.5% sodium deoxycholate) with proteinase inhibitors added [0.1 mM orthovanadate/0.1 mM phenylmethylsulfonyl fluoride/leupeptin (5 μg/ml)/aprotinin (1 μg/ml)]. Total protein concentration of cell extracts was determined by the Bradford microassay. Protein extracts (80 μg) were solublized in 0.2 vol of 5× concentrated sample buffer (0.25 M Tris⋅HCl, pH 6.8/0.4 M dithiothreitol/5% SDS/0.5% bromophenol blue/10% glycerol) and separated on 5% low-cross-linked polyacrylamide gel (monomer to crosslinker ratio, 100:1) for ATM and a 12% gel for WAF1. After electrophoresis, proteins were transferred to poly(vinylidene difluoride) membranes (DuPont) as described previously (35). The filters were blotted in 5% skim milk overnight, then probed with ATM or WAF1 antibodies, and visualized by using the ECL method (Amersham). Densitometric scanning was used to quantify protein. Recombinant ATM protein was immunoprecipitated from lysates of transfected cells with anti-hexahistidine antibody (CLONTECH) and protein G-Sepharose overnight at 4°C. Resulting protein complexes were washed in lysis buffer and electrophoresed on 5% SDS/PAGE gels as described above.
Purification of Recombinant ATM Protein.
The recombinant ATM protein expressed from pMAT1, which carries a 6×His tag at its N terminus, was purified using a nickle resin (CLONTECH). Stably transfected logarithmically growing A-T cells were pelleted, washed once with ice-cold PBS, and lysed with RIPA buffer with proteinase inhibitors added. The mixture was sonicated (two 10-sec periods). After incubation on ice for 30 min, the lysate was centrifugated at 10,000 × g for 15 min. A volume of 50 μl of nickle chelate resin was added to 2 mg of the cleared protein extract, and the mixture was rotated at 4°C for 1 h and centrifuged at 2,000 × g for 2 min. The pellet was washed with RIPA buffer three times and the bound ATM-(His) protein was eluted with RIPA buffer containing 50 mM imidazole. The eluted protein samples were solublized in SDS/protein sample buffer at 37°C for 20 min and analyzed by Western blot analysis.
[3H]Thymidine Incorporation Assay.
Logarithmically growing cells transfected with pMAT1 were incubated at 37°C for 48 h in RPMI 1640 medium containing [14C]thymidine (0.02 μCi/ml; 1 Ci = 37 GBq). The cells were washed twice with PBS, resuspended in RPMI 1640 medium, and incubated at 37°C for 2 h. A volume of 75 μl of cells (4 × 104 cells) was mixed in a well of a 96-well microtiter plate with 75 μl of complete medium with or without CdCl2 (5 μM). After incubation for 16 h at 37°C, the cells were irradiated with 0, 5, 10, 20, or 30 Gy of γ-radiation. [3H]Thymidine (Dupont-NEN, NET027X) (0.2 μCi/ml) was used to measure DNA synthesis for 3 h at 37°C after irradiation.
SAPK/JNK Assay.
Glutathione S-transferase (GST)–Jun (residues 1–79) was used as a substrate to measure activation of SAPK. Cells were irradiated with 20 Gy of radiation and incubated for 1 h. Cells were lysed in universal immunoprecipitation buffer, and total cell lysate was immunoprecipitated with anti-SAPK antibody (SAPK β, Santa Cruz Biotechnology). Immune complex kinase assays were preformed by incubating the resulting protein complexes in kinase buffer (25 mM Tris⋅HCl, pH 7.4/10 mM MgCl2/1 mM MnCl2/0.5 mM dithiothreitol/10 μM ATP), with 5 μg of GST–Jun and 5 μCi of [γ-32P]ATP for 30 min at 20°C, and analyzed by SDS/PAGE on 12% gels.
RESULTS AND DISCUSSION
Based on the complete sequence of the ATM ORF, obtained from cDNA contigs (32), we designed a set of oligonucleotide primers to amplify a 4.7-kb cDNA fragment corresponding to the 5′ end of the ATM mRNA sequence, prepared from normal lymphoblastoid cells. The 4.7-kb PCR fragment extends from immediately downstream of the ATG initiation codon of ATM and overlaps with the 5.9-kb partial cDNA (Fig. 1). Both cDNA fragments were used to construct a full-length cDNA clone as outlined in Fig. 1. This cDNA was cloned into an EBV-based vector containing a hexahistidine tag (pEBV-His-B) located downstream and in-frame of an ATG initiation codon contained in the vector. This construct, pEAT11, constitutively expresses the ATM protein under the control of an Rous sarcoma virus promoter. The hexahistidine tag-ATM cDNA was subcloned into an EBV-based vector pMEP4 to create pMAT1, allowing inducible expression of ATM cDNA directed by a metallothionein II promoter and induced by CdCl2 (33). The advantage of this construct is that a conditional high-level expression of ATM can be achieved since it was possible that endogenous mutated ATM might interfere with basal levels of expression of the transfected cDNA. It should be pointed out that the 5′ end of the ORF is unstable when propagated in Escherichia coli. In most of the clones with inserts, deletion had occurred at the 5′ end with as many as 460 nucleotides being deleted (results not shown). To circumvent problems associated with the unstable sequence at the 5′ end of the gene, the clones were screened with probe against the extreme end of the 5′ ORF, and approximately 1–2% of the clones contained the full-length cDNA. DNA sequence analysis was carried out and the integrity of the ATM cDNA was confirmed by comparison with the ATM cDNA contig sequence in GenBank (accession no. U33841). The sequence obtained differed by only three nucleotides from that deposited, two of these changes did not alter the amino acid and the third ATG to ATA (codon 847) resulted in the substitution of isoleucine for methionine, both of which contain nonpolar side chains. It is likely that this is a polymorphism since we demonstrate in this report that this cDNA is capable of correcting the A-T phenotype. Furthermore, only one missense mutation has been described in this gene in more than 100 A-T cell lines, and that mutation is located within the phosphatidylinositol-3-kinase domain (36). All of the other mutations to date are predicted to give rise to protein truncations or involve small-to-large in-frame deletions (22, 37, 38)
Figure 1.
Isolation and cloning of full-length ATM cDNA. The strategy involved combining a partial 5.9-kb ATM cDNA (22) with an overlapping 4.7-kb PCR fragment from the 5′ end of the mRNA to generate a fragment of 9.58 kb. This cDNA was cloned into an EBV-based vector (pEBV-His-B) containing a hexahistidine tag sequence to generate pEAT11 for constitutive expression. The insert and the hexahistidine sequence from this construct was subsequently subcloned into a second EBV-vector pMEP4 (33), which contains a metallothionein II inducible promoter. This construct was designated pMAT1.
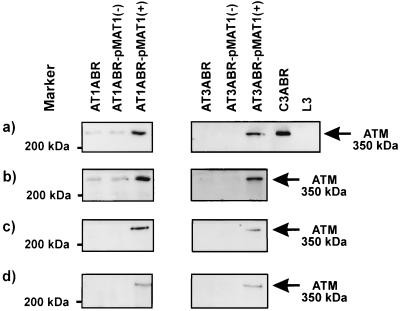
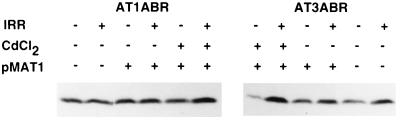
It has been demonstrated that A-T heterozygotes show intermediate sensitivity to radiation, as judged by a number of different parameters (4, 39–43), which may be due to interference from the product of the mutated allele. Accordingly, we studied the effects of ATM expression in A-T cells by using the inducible construct pMAT1 to produce significant amounts of the ATM protein. Two A-T cell lines AT1ABR (homozygous 9-bp in-frame deletion, capable of producing near full-length protein) and AT3ABR (compound heterozygote for mutations giving rise to truncated proteins) (22, 44) were selected for transfection with pMAT1. Selection was carried out with hygromycin for 4 weeks and stably transfected cells were selected for further studies. The presence of episomal plasmid DNA was determined by Southern blot analysis, revealing that the different transfected cell lines had approximately equal copy numbers of pMAT1 (results not shown). Restriction enzyme mapping confirmed that there were no deletions or rearrangements in the ATM insert. ATM protein expression was determined by immunoblotting cell extracts prepared after CdCl2 treatment (5 μM for 16 h), which induced transcription of the ATM gene from the metallothionein MTII promoter in pMAT1. As expected, ATM protein was detected by immunoblotting with ATM-1.8 polyclonal antibody (prepared against a recombinant C-terminal fragment of ATM) in transfected uninduced AT1ABR cells (Fig. 2a). There was a significant increase in the amount of ATM protein after induction with CdCl2 (Fig. 2a). No endogenous ATM protein was detected in uninduced AT3ABR cells, as expected, but again induction with CdCl2 led to an increase in ATM protein to a level similar to that in AT1ABR (Fig. 2a). Similar results were observed with a second polyclonal antibody, ATM-4BA, also raised against a recombinant protein fragment corresponding to a C-terminal (amino acids 2323–2740) portion of the molecule (Fig. 2b and ref. 44). The faint band detected by ATM-4BA antibody in uninduced AT3ABR cells appears to be due to cross-reaction to an unrelated protein since mutations in both ATM alleles are predicted to give rise to truncated proteins that would terminate upstream of the antibody recognition site (44). Furthermore, a third polyclonal antibody, ATM-3BA (an anti-peptide antibody against amino acids 2581–2599) also failed to detect ATM protein in this cell line (44). Since the expressed recombinant ATM contains a hexahistidine tag at the N terminus, it was possible to purify ATM with a nickel chelate resin. As expected, immunoblotting of this material revealed the presence of recombinant protein only in CdCl2-induced cells (Fig. 2c). On the basis of this property, it was also possible to detect induced ATM by using immunoprecipitation with an anti-hexahistidine antibody (Fig. 2d).
Figure 2.
Expression of full-length ATM cDNA in A-T cells. EBV-transformed lymphoblastoid cells were transfected with pMAT1 by using the Lipofectin reagent (Life Technologies) according to the manufacturer’s instructions. Stably transfected A-T cells were induced with 5 μM CdCl2 (+) for 16 h or were uninduced (−). Total cell extract from pMAT1 transfected AT1ABR and AT3ABR were immunoblotted with different ATM antibodies. A control cell line, C3ABR, and an A-T cell line, L3 (homozygous mutations at nucleotide 120 predicted to give rise to truncated protein 35 amino acids long), are also included. (a) ATM-1.8 antibodies. (b) ATM4BA antibodies. (c) Analysis of samples purified from cell extracts by using a nickle chelate resin to isolate His-tagged protein. Detection was carried out with ATM4BA antibodies. (d) Analysis of samples immunoprecipitated with anti-hexahistidine antibody (CLONTECH) followed by immunoblotting with ATM4BA antibody.
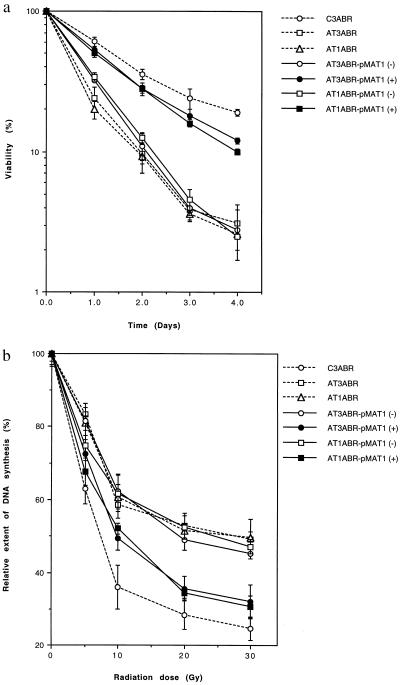
A universal characteristic of A-T is hypersensitivity to radiation that is manifested by enhanced cell killing and higher levels of induced chromosome aberrations (3, 4, 45). This characteristic was initially selected to check for correction of the radio-sensitive phenotype by ATM cDNA. Cell viability was determined by trypan blue exclusion, which we have shown previously reflects accurately the survival difference between control and A-T cells after exposure to radiation (4). As expected, the kinetics of cell survival in pMAT1 transfected uninduced AT1ABR and AT3ABR cells (exposed to 4 Gy of radiation) was comparable to that for untransfected AT1ABR and AT3ABR but was much more sensitive than for a control cell line, C3ABR (Fig. 3a). In contrast when AT1ABR and AT3ABR cells were exposed to radiation after CdCl2 induction, cell survival increased to a level similar to the control cell line (Fig. 3a). Similar observations were made at other radiation doses in the range 1–4 Gy (results not shown). Transfection with pEAT11, which constitutively expresses ATM, also conferred resistance to wild-type AT1ABR and AT3ABR cells (results not shown).
Figure 3.
Correction of the radio-sensitive phenotype in A-T cells. (a) Cell survival. pMAT1 transfected AT1ABR and AT3ABR cells were induced (+) or mock-induced (−) for ATM expression with 5 μM CdCl2 for 16 h. Cell viability was determined as described (4). Nontransfected A-T and normal cells were used as controls. Each point represents an average of triplicate experiments. Bars are SD. (b) Complementation of radio-resistant DNA synthesis in A-T cells after transfection with pMAT1. CdCl2 treated (+) and untreated (−) are shown. Prelabeled cells were induced with CdCl2 for 16 h and irradiated, and DNA synthesis was determined by labeling with [3H]thymidine (0.2 μCi/ml) for 3 h. DNA synthesis is expressed relative to that in unirradiated cells. Points represent mean ratios of thymidine incorporation.
Induced chromosome aberrations are also a good indicator of radio-sensitivity in A-T (2, 20). The results in Table 1 reveal that CdCl2 induction of recombinant ATM protein in transfected AT1ABR and AT3ABR cells led to a reduction in radiation-induced chromosome aberrations to levels comparable to those in irradiated control cells. This correction was not observed in uninduced cells since aberrations were of the same order as those in untransfected A-T cells, approximately 3-fold higher than in controls (Table 1). A similar observation was made with the constitutively expressing construct pEAT11 (results not shown).
Table 1.
Correction of γ-radiation-induced G2-phase chromosome aberrations in A-T cells by using full-length ATM cDNA cloned into the inducible vector pMAT1
| Cell line | Aberrations, no.
|
No. ICA/ metaphase | ||
|---|---|---|---|---|
| sb | cb | Int | ||
| C3ABR | 57 | 0 | 0 | 1.14 |
| AT1ABR | 145 | 0 | 1 | 2.92 |
| AT1ABR + pMAT1 | 150 | 0 | 0 | 3.00 |
| AT1ABR + pMAT1 (induced) | 62 | 2 | 0 | 1.24 |
| AT3ABR | 140 | 1 | 2 | 2.86 |
| AT3ABR + pMAT1 | 144 | 0 | 0 | 2.88 |
| AT3ABR + pMAT1 (induced) | 58 | 1 | 0 | 1.18 |
sb, Chromatid breaks; cb, chromosome breaks; Int, interchanges; ICA, induced chromosome aberrations. Fifty metaphases were analyzed for each sample after exposure to 1 Gy of radiation. AT1ABR and AT3ABR are untransfected cells. Data for ICA are the mean.
Radio-resistant DNA synthesis is also a universal characteristic of A-T cells (4). Transfection with ATM cDNA would be predicted to restore a normal pattern of inhibition of DNA synthesis to A-T cells after irradiation. As is evident in Fig. 3b, DNA synthesis is inhibited in a biphasic pattern in control (C3ABR) cells with increasing radiation dose, but the extent of inhibition is markedly less in AT1ABR and AT3ABR cells, as well as in their uninduced transfected counterparts. However, after induction of ATM expression in AT1ABR and AT3ABR cells, radiation exposure resulted in a significant shift in the kinetics of DNA synthesis inhibition, similar to that observed in irradiated control cells (Fig. 3b).
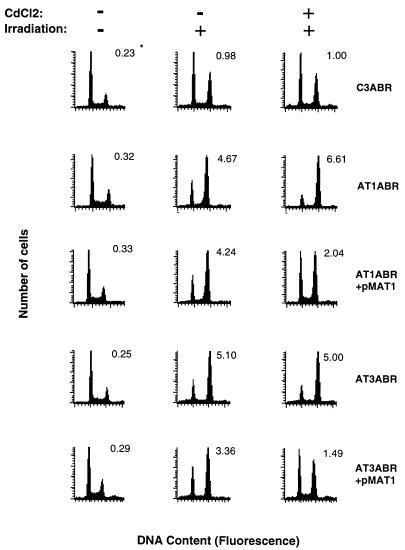
In addition to the defect in the S-phase checkpoint (radio-resistant DNA synthesis), A-T cells are also defective in activating both the G1- and G2-phase checkpoints shortly after irradiation (5, 6), and at longer times (15–24 h) after irradiation A-T cells are irreversibly blocked at G2/M phase, where they die (7, 46, 47). It is evident from the results presented in Fig. 4 that the accumulation of A-T cells blocked at G2/M phase 24 h after irradiation (AT1ABR, G2/G1 = 6.61; AT3ABR, G2/G1 = 5.00) is significantly greater than in control cells (G2/G1 = 1.00). Comparable results were obtained with transfected uninduced A-T cells but the extent of G2 delay was greatly reduced in CdCl2-induced ATM-expressing AT1ABR (G2/G1 = 2.04) and AT3ABR (G2/G1 = 1.49) cells.
Figure 4.
Extent of G2-phase delay at 24 h after irradiation. Transfected and untransfected cells were treated with 5 μM CdCl2 16 h prior to irradiation. Cells were irradiated (3 Gy), incubated for 24 h, harvested, and analyzed for DNA content by flow cytometry as described (7). *Values represent the ratio of G2- to G1-phase cell numbers, which was chosen as a more exact representation of delay in G2 phase (48).
Failure of A-T cells to activate the G1 checkpoint appears to be due to a defect in the radiation signal transduction pathway operating through p53, its target gene, CDKN1A encoding p21/WAF1 and cyclin-dependent kinases (14–16). The mitotic inhibitor nocodazole was used to provide an accurate measurement of exit from G1 phase by blocking cells leaking past the G2 checkpoint from reentering G1 phase (49). As observed previously (7), the percentage of C3ABR cells delayed in G1 phase (36.0 ± 3.5%) was considerably greater than that for either of the two A-T cell lines (7–8%), and CdCl2 treatment did not alter this significantly (Table 2). However, a partial correction of the G1 checkpoint was achieved in pMAT1-transfected AT1ABR and AT3ABR cells after induction of ATM expression with CdCl2, with the proportion of cells delayed in G1 phase increased to 18.1 ± 0.3% and 18.5 ± 1.3%, respectively, compared with a value of 33.8 ± 3.7% for the control (Table 2). As a further indicator of the restored integrity of the G1 checkpoint, we determined the radiation induction of p21/WAF1, which has been shown to be defective in A-T cells (14–16). The results in Fig. 5 demonstrate that the ATM cDNA restored the induction of p21/WAF1 only in CdCl2-induced extracts from pMAT1-transfected AT1ABR and AT3ABR.
Table 2.
Partial correction of delay of cells in G1-phase after irradiation
| CdCl2 | % cells in G1 phase
|
|
|---|---|---|
| −CdCl2 | +CdCl2 | |
| C3ABR | 36.0 ± 3.5 | 33.8 ± 3.7 |
| AT1ABR | 7.8 ± 0.8 | 6.3 ± 1.1 |
| AT1ABR + pMATI | 8.2 ± 0.4 | 18.1 ± 0.3 |
| AT3ABR | 6.8 ± 0.7 | 6.3 ± 0.4 |
| AT3ABR + pMATI | 6.7 ± 0.4 | 18.5 ± 1.3 |
Cells were treated (+) or mock-treated (−) with CdCl2 (5 μM) for 16 h, irradiated (3 Gy), and incubated in the presence of nocodazale (2 mM) for 24 h. Percentage of cells accumulating in G1 phase after irradiation was determined by flow cytometric analysis. Data represent the mean ±SD of two experiments.
Figure 5.
Induction of p21/WAF1 after exposure of cells to radiation. Cells were exposed to radiation (3 Gy) after prior incubation with CdCl2 for 16 h or without CdCl2 treatment, and p21/WAF1 was determined by immunoblotting with anti-WAF1 antibody (OP-64, Oncogene Science). Protein loading was assessed by staining membranes with Poinceau S.
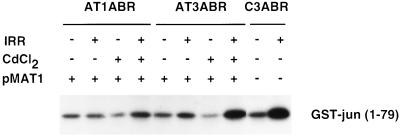
A-T cells are also defective in the induction of stress-activated protein kinase (SAPK/JNK) by ionizing radiation (50). To determine whether ATM cDNA corrected this defect, we assayed for radiation activation of SAPK by immunoprecipitating cell lysates with anti-JNKI/SAPKβ antibody and determining the extent of phosphorylation of a GST–Jun substrate (residues 1–79) in vitro. SAPK activity increased 5-fold in control cells in response to radiation (20 Gy), but there was no significant change in either of the uninduced A-T cell lines (Fig. 6). On the other hand after CdCl2 treatment, pMAT1-transfected AT3ABR cells had a normal SAPK response, whereas transfected AT1ABR were intermediate in their response (Fig. 6). A possible explanation for this is the presence of near full-length protein in AT1ABR cells, which may have more of an effect on specific aspects of the phenotype.
Figure 6.
Activation of SAPK by ionizing radiation. Cells were treated with 20 Gy of ionizing radiation and harvested 1 h later. Total lysates were immunoprecipitated with anti-SAPK antibody (SAPK β, Santa Cruz Biotechnology), and in vitro immune complex kinase assays were performed with GST–Jun (residues 1–79) as substrate.
The large size of the ATM protein (350 kDa) and the complex phenotype in A-T suggest that ATM has more than one function. Since the phosphatidylinositol-3-kinase domain occupies only a small part of ATM, it is likely that other domains are also important for the function of the ATM protein. A partial leucine-zipper motif has been identified (amino acids 1217–1238) and we have recently demonstrated the importance of a sequence, DPAPNPPHFP (amino acids 1373–1382), in the binding of the ATM protein to the protein-tyrosine kinase c-Abl (51), an important factor in the stress response to ionizing radiation (52). This protein kinase is required for the activation of SAPK after irradiation (52), a response that is defective in A-T (50, 51). Correction of the SAPK defect in A-T cells by ATM cDNA suggests strongly that the ATM–cAbl interaction is necessary for the DNA damage response, but it remains unclear at which stage of the cell cycle this signal acts.
Our results demonstrate that full-length ATM cDNA corrects the radio-sensitive phenotype in A-T cells. This was achieved under conditions where overexpression of ATM was employed. It is also evident that in some of the assays used to screen for correction of the defective phenotype in A-T, this correction was not completely effective, even when the ATM product was overexpressed. This can be explained either by the complex phenotype in A-T (1) or due to a dominant negative effect from the mutated form of endogenous ATM. This is supported by the observation that some A-T heterozygotes show intermediate response to radiation (4, 39–43). By using this approach, it should be possible to determine whether specific defects in A-T cells are correlated with or dependent upon the expression of particular ATM protein regions, and this should assist in the identification of potential functional domains in ATM. The correction of multiple cellular A-T defects by expression of ATM cDNA enhances the potential for correction of both the neurological and immunological abnormalities in A-T by gene therapy with this ATM cDNA. Furthermore, the expression of antisense ATM cDNA, using the appropriate delivery systems, should also provide a new approach to sensitizing tumors to radiation and thereby benefit tumor therapy.
Acknowledgments
We thank Nicholas Hayward and Ann Tresize for critically reading the manuscript and Ann Knight for typing. We are also grateful to Aine Farrell and Karen Hobson for assistance with cell culture and Bernadette Garrone for help with photography. We thank the National Health and Medical Research Council of Australia, the Queensland Cancer Fund, the A-T Childrens Project, and the Tzu Chi Foundation for support. N.Z. was supported by a grant from the National Institutes of Health, Bethesda, MD.
ABBREVIATIONS
- A-T
ataxia-telangiectasia
- ATM
A-T mutated
- EBV
Epstein–Barr virus
- GST
glutathione S-transferase
References
- 1.Sedgwick R P, Boder E. In: Hereditary Neuropathies and Spinocerebellar Atrophies. Viannet De Jong J M B, editor. Amsterdam: Elsevier; 1991. pp. 347–423. [Google Scholar]
- 2.Aurias A B, Dutrillaux D B, Lejeune J. Mutat Res. 1980;69:369–374. doi: 10.1016/0027-5107(80)90101-3. [DOI] [PubMed] [Google Scholar]
- 3.Taylor A M R, Harnden D G, Arlett C F, Harcourt S A, Lehmann A R, Stevens S, Bridges B A. Nature (London) 1975;4:427–429. doi: 10.1038/258427a0. [DOI] [PubMed] [Google Scholar]
- 4.Chen P C, Lavin M F, Kidson C, Moss D. Nature (London) 1978;274:484–486. doi: 10.1038/274484a0. [DOI] [PubMed] [Google Scholar]
- 5.Houldsworth J, Lavin M F. Nucleic Acids Res. 1980;8:3709–3720. doi: 10.1093/nar/8.16.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Painter R B, Young B R. Proc Natl Acad Sci USA. 1980;77:7315–7317. doi: 10.1073/pnas.77.12.7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beamish H, Lavin M F. Int J Radiat Biol. 1994;65:175–184. doi: 10.1080/09553009414550211. [DOI] [PubMed] [Google Scholar]
- 8.Zampetti-Bosseler F, Scott D. Int J Radiat Biol. 1981;39:547–558. doi: 10.1080/09553008114550651. [DOI] [PubMed] [Google Scholar]
- 9.Kastan M B, Zhan O, EL-, Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 10.Khanna K K, Lavin M F. Oncogene. 1993;8:3307–3312. [PubMed] [Google Scholar]
- 11.Lu X, Lane D P. Cell. 1993;75:765–778. doi: 10.1016/0092-8674(93)90496-d. [DOI] [PubMed] [Google Scholar]
- 12.Artuso M, Esteve A, Bresil H, Vuillaume M, Hall J. Oncogene. 1995;8:1427–1435. [PubMed] [Google Scholar]
- 13.Mirzayans R, Famulski K S, Enns L, Fraser M, Paterson M C. Oncogene. 1995;8:1597–1605. [PubMed] [Google Scholar]
- 14.Khanna K K, Beamish H, Yan J, Hobson K, Williams R, Dunn I, Lavin M F. Oncogene. 1995;11:609–618. [PubMed] [Google Scholar]
- 15.Canman C E, Wolff A C, Chen C Y, Fornace A J, Jr, Kastan M B. Cancer Res. 1994;54:5054–5058. [PubMed] [Google Scholar]
- 16.Beamish H, Williams R, Chen P, Khanna K K, Hobson K, Watters D, Shiloh Y, Lavin M F. J Biol Chem. 1996;271:20486–20493. doi: 10.1074/jbc.271.34.20486. [DOI] [PubMed] [Google Scholar]
- 17.Meyn M S. Cancer Res. 1995;55:5591–6001. [PubMed] [Google Scholar]
- 18.Ziv Y, Bar-Shira A, Jorgensen T J, Russell P S, Sartiel A, Shows T B, Eddy R L, Buchwald M, Legerski R, Schimke R T, Shiloh Y. Somatic Cell Mol Genet. 1995;21:99–111. doi: 10.1007/BF02255785. [DOI] [PubMed] [Google Scholar]
- 19.Jung M, Zhang Y, Lee S, Dritschilo A. Science. 1995;268:1619–1621. doi: 10.1126/science.7777860. [DOI] [PubMed] [Google Scholar]
- 20.Chen P, Girges A A, Hobson K, Beamish H, Khanna K K, Farrell A, Gatei M, Teale B, Buchwald M, Legerski R, Lavin M F. Int J Radiat Biol. 1996;69:385–395. doi: 10.1080/095530096145940. [DOI] [PubMed] [Google Scholar]
- 21.Gatti R, Berkel I, Boder E, Braedt G, Charmley P, et al. Nature (London) 1988;336:577–580. doi: 10.1038/336577a0. [DOI] [PubMed] [Google Scholar]
- 22.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, et al. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 23.Uziel T, Savitsky K, Platzer M, Ziv Y, Helbitz T, Nehls M, Boehm T, Rosenthal A, Shiloh Y, Rotman G. Genomics. 1996;33:317–320. doi: 10.1006/geno.1996.0201. [DOI] [PubMed] [Google Scholar]
- 24.Jackson S P. Curr Biol. 1995;5:1210–1212. doi: 10.1016/s0960-9822(95)00238-7. [DOI] [PubMed] [Google Scholar]
- 25.Lavin M F, Khanna K K, Beamish H, Williams R, Spring K, Watters D, Shiloh Y. Trends Biochem Sci. 1995;20:382–383. doi: 10.1016/s0968-0004(00)89083-0. [DOI] [PubMed] [Google Scholar]
- 26.Zakian V A. Cell. 1995;82:685–687. doi: 10.1016/0092-8674(95)90463-8. [DOI] [PubMed] [Google Scholar]
- 27.Hari K L, Santerre A, Sekelsky J J, McKim K S, Boyd J B, Hawley R S. Cell. 1995;82:815–821. doi: 10.1016/0092-8674(95)90478-6. [DOI] [PubMed] [Google Scholar]
- 28.Hartley K O, Gell D, Smith G C M, Zhang H, Divecha N, Connelly M A, Admon A, Lees-Miller S P, Anderson C W, Jackson S P. Cell. 1995;82:849–856. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- 29.Morrow D M, Tagle D A, Shiloh Y, Collins F S, Hieter P. Cell. 1995;82:831–840. doi: 10.1016/0092-8674(95)90480-8. [DOI] [PubMed] [Google Scholar]
- 30.Paulovich A G, Hartwell L H. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- 31.Greenwell P W, Kronmal S L, Porter S E, Gassenhuber J, Obermaier B, Petes T D. Cell. 1995;82:823–829. doi: 10.1016/0092-8674(95)90479-4. [DOI] [PubMed] [Google Scholar]
- 32.Savitsky K, Sfez S, Tagle D, Ziv Y, Sartiel A, Collins F S, Shiloh Y, Rotman G. Hum Mol Genet. 1995;4:2025–2032. doi: 10.1093/hmg/4.11.2025. [DOI] [PubMed] [Google Scholar]
- 33.Vousden K H, Crook T, Farrell P H. J Gen Virol. 1993;74:803–810. doi: 10.1099/0022-1317-74-5-803. [DOI] [PubMed] [Google Scholar]
- 34.Strathdee C A, Gavish H, Shannon W R, Buchwald M. Nature (London) 1992;356:763–767. doi: 10.1038/356763a0. [DOI] [PubMed] [Google Scholar]
- 35.Song Q, Lees-Miller S P, Kumar S, Zhang N, Chan D W, Smith G C M, Jackson S P, Alnemri E S, Litwack G, Khanna K K, Lavin M F. EMBO J. 1996;15:3238–3246. [PMC free article] [PubMed] [Google Scholar]
- 36.Gilad S, Khosravi R,, Shkedy D,, Uziel T,, Ziv Y, et al. Hum Mol Genet. 1996;5:433–440. doi: 10.1093/hmg/5.4.433. [DOI] [PubMed] [Google Scholar]
- 37.Byrd P J, McConville C M, Cooper P, Parkhill J, Stankovic T, McGuire G M, Thick J A, Taylor A M R. Hum Mol Genet. 1996;5:145–149. doi: 10.1093/hmg/5.1.145. [DOI] [PubMed] [Google Scholar]
- 38.Telatar M, Wang Z, Udar W, Liang T, Concannon P, Bernatowska-Matuscklewicz E, Lavin M F, Sholoh Y, Good R A, Gatti R A. Am J Hum Genet. 1996;59:40–44. [PMC free article] [PubMed] [Google Scholar]
- 39.Paterson M C, Anderson A K, Smith B P, Smith P J. Cancer Res. 1979;39:3725–3734. [PubMed] [Google Scholar]
- 40.Cole J, Arlett C F, Green M H L, Harcourt S A, Priestley A, Henderson L, Cole H, James E, Richmond F. Int J Radiat Biol. 1988;54:929–942. doi: 10.1080/09553008814552331. [DOI] [PubMed] [Google Scholar]
- 41.Waghray M, Al-Sedairy S, Ozand P T, Hannan M A. Hum Genet. 1990;84:532–534. doi: 10.1007/BF00210804. [DOI] [PubMed] [Google Scholar]
- 42.Rosin M P, Ochs H D. Hum Genet. 1986;74:335–340. doi: 10.1007/BF00280482. [DOI] [PubMed] [Google Scholar]
- 43.Sanford K K, Parshad R, Price F M, Jones G M, Tarone R E, Eirman L, Hale P, Waldmann T. J Natl Cancer Inst. 1990;82:1050–1054. doi: 10.1093/jnci/82.12.1050. [DOI] [PubMed] [Google Scholar]
- 44.Watters D, Khanna K K, Beamish H, Birrell G, Spring K, Kedar P, Gatei M, Stenzel D, Hobson K, Kozlov S, Farrell A, Ramsay J, Gatti R, Lavin M F. Oncogene. 1997;14:1911–1921. doi: 10.1038/sj.onc.1201037. [DOI] [PubMed] [Google Scholar]
- 45.Lavin M F, Shiloh Y. Annu Rev Immunol. 1997;15:177–202. doi: 10.1146/annurev.immunol.15.1.177. [DOI] [PubMed] [Google Scholar]
- 46.Ford M D, Martin L, Lavin M F. Mutat Res. 1984;125:115–122. doi: 10.1016/0027-5107(84)90038-1. [DOI] [PubMed] [Google Scholar]
- 47.Bates P R, Imray F P, Lavin M F. Int J Radiat Biol. 1985;47:713–722. doi: 10.1080/09553008514550951. [DOI] [PubMed] [Google Scholar]
- 49.Fan S, El-Deiry W S, Bae I, Freeman J, Jondle D, Bhatia K, Fornace A J, Jr, Magrath I, Kohn K W, O’Connor P M. Cancer Res. 1994;54:5824–5830. [PubMed] [Google Scholar]
- 50.Shafman T D, Saleem A, Kyriakis J, Weichselbaum R, Kharbanda S, Kufe D W. Cancer Res. 1995;55:3242–3245. [PubMed] [Google Scholar]
- 51.Khanna K K, Shafman T D, Kedar P, Spring K, Kozlov S, Yen T, Gatei M, Zhang N, Watters D, Egerton M, Shiloh Y, Lavin M F. Nature (London) 1996;387:520–524. doi: 10.1038/387520a0. [DOI] [PubMed] [Google Scholar]
- 52.Kharbanda S, Ren R, Pandey P, Shafman T D, Feller S M, Weichselbaum R R, Kufe D W. Nature (London) 1995;376:785–788. doi: 10.1038/376785a0. [DOI] [PubMed] [Google Scholar]
- 48.Lavin M F, Bennett I, Ramsay J, Gardiner R A, Seymour G J, Farrell A, Walsh M. J Natl Cancer Inst. 1994;86:1627–1634. doi: 10.1093/jnci/86.21.1627. [DOI] [PubMed] [Google Scholar]