Abstract
In mammals, at least, a species-specific mechanism exists that eliminates sperm-derived mitochondrial DNA from a fertilized egg. The result is the “one female ancestor per generation” rule and three other rules of mitochondrial inheritance. The second, third, and fourth rules are as follows. (ii) Sublineages of a given mitochondrial line can be generated only during the parallel descents from ancestral sisters. (iii) In a static population in which the production of one female progeny per mated pair per generation has been a rule, several ancient mitochondrial lineages harking back to the female founders of the speciation may persist side by side. (iv) Two or more individuals not related to each other in the recent past may share the identical or nearly identical mitochondrial genome derived from the common female ancestor or ancestral sisters of many generations ago.
Compared with the great deal of attention that traditionally has been paid to the problem of descents, the problem of ascents has largely been ignored. In an earlier paper, the three cardinal rules of ancestry were set forth: (i) the law of sibling interference, which progressively curtails exponential increases in numbers of one’s ancestors; (ii) the presence of the past ancestry-saturated generation during which nearly all progeny-produced adults of a population became one’s ancestors; and (iii) the law of increasingly irrelevant remote ancestors. Delving into the remote past reveals that smaller and smaller fractions of ancestors left their genetic traces in diploid genomes (1). The present paper reveals the four cardinal rules of mitochondrial inheritance.
A number of cytoplasmic organelles of eukaryotes were derived from endosymbiotic bacteria (2). Of these, the mitochondrion still retains a circular genomic DNA of its own, albeit in an extremely abbreviated form. The mitochondrion is thought to be derived from the aerobic, purple, nonsulfur eubacterium resembling Paracoccus denitrificans of today (3). It has been shown that in interspecific hybrids between rodents Mus musculus and Mus spretus, the sperm-derived mitochondrial DNA coexists with its egg-derived counterpart throughout embryonic and fetal development. In the case of intraspecific crosses, on the contrary, the sperm-derived mitochondrial DNA disappears all together almost immediately after the fertilization within the pronucleus stage (4). The above has revealed the presence of a species-specific elimination mechanism that ensures the strictly maternal inheritance of the mitochondrial genome within each mammalian species.
The One Ancestor per Generation Rule of Mitochondrial Inheritance
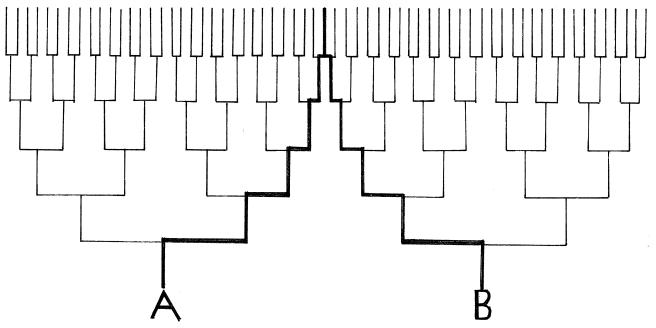
The strictly maternal inheritance of mitochondrial genomes due to the selective elimination of sperm-derived mitochondrial DNA implies that there can never be recombinations (mixing) between mitochondrial genomes. It follows that so far as the mitochondrial genome is concerned, each is destined to become an uncontaminatable lineage of its own. Accordingly, each individual could have had only one ancestor at each ancestral generation, no matter how far back one traces one’s ancestry. This principle is illustrated in Fig. 1 (Left and Right) with regard to ancestors of individuals A and B up to five generations back.
Figure 1.
Combined genealogical trees up to five generations back of individuals A and B. In the genealogical tree of A, a female ancestor is always placed to the right of a male ancestor, whereas converse placements are followed in that of B. The common mitochondrial ancestry of the two are indicated by thick lines. This figure simultaneously illustrates the first and fourth rules of mitochondrial inheritance.
The Second Rule: Only Ancestral Sisters Generate Sublineages of the Stem Mitochondrial Genome
The most mutable part of the circular mitochondrial genome is the D (displacement)-loop region, which is noncoding. Furthermore, in the case of human mitochondria, two hypervariable segments, each roughly 400-bp long, were detected within this D-loop region. Wilson and coworkers (5) performed the seminal study on these two hypervariable segments of mitochondrial genomes derived from diverse human individuals. They established that the maximal sequence divergence observed among sampled human individuals of varied origins is only 2%, differences represented exclusively by transitional base substitutions. This revealed the very recent origin of our own species, Homo sapiens. In fact, our own species resembled a subspecies rather than the whole of the well established species as to the degree of sequence divergence. For example, the sequence divergence among the comparable mitochondrial segments of the red junglefowl Gallus gallus was 7%, and base substitutions were not only transitions but also more drastic transversions (6). The divergence comparable to that of our own species was found in each of the three subspecies of the red jungle fowl (e.g., Gallus gallus spadiceus) (6).
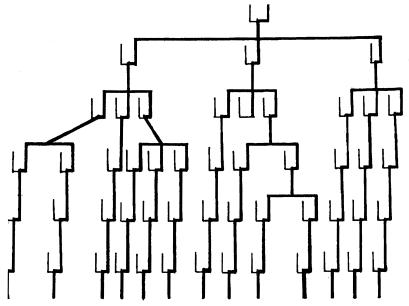
The seminal study on human mitochondria also showed that African sequences were far more diverse than those of Asians and Europeans combined, thus confirming the African origin of our own species (5). The dendrogram of deduced human mitochondrial lineages shown in figure 3 of the seminal study clearly revealed the presence not only of ancient stem mitochondrial lineages but also the diversification of many of these stem lineages to sublineages (5). In the case of 12 Western Pygmies from Africa, the presence of three stem mitochondrial lineages differing from each other by as much as 1.4% (11 base substitutions) was quite evident. Each of these three stem lineages, however, split into a number of sublineages. For example, mitochondrial genomes of four Pygmies clearly belonged to one of the stem lineages. Yet, these four individual mitochondrial genomes differed from each other by two base substitutions each (0.25% difference). Such a diversification into a number of sublineages can readily be explained by the presence of maternal sisters in their common ancestry, as illustrated in Fig. 2. Originally, a set of full or maternal half-sisters should have inherited the identical mitochondrial genome from their mother. However, because each sister’s female descendants invariably establish an independent lineage (or lineages), in time it would accumulate its own characteristic mutations to become a distinct sublineage. Inasmuch as the substitution rate for the human D-loop region has since been estimated as 7 × 10−8/site per year (7), a great deal more than 10,000 years (500 generations) of the independent existence is required before these sister-derived mitochondrial sublineages become distinct, differing from each other by at least single base substitutions.
Figure 2.
Ancestral sisters as generators of mitochondrial sublineages are schematically illustrated. Mitochondrial (maternal) lineages are traced in thick, black lines, and a male consort of each mating is indicated by a short, thin line. The possessor of one stem-mitochondrial lineage at the top produced three daughters, and each of the three daughters, in turn, produced three granddaughters, thus initiating the diversification of the stem lineage into 13 sublineages.
The Third Rule: Eight Eves for People of the Kung Tribe in Africa
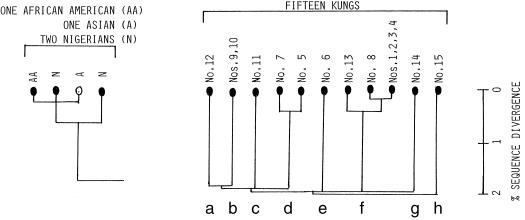
As already noted in an earlier paper (1), large-bodied omnivorous animals such as bears consume prodigious amounts of flesh, grains, roots, and fruits and contaminate the environment with proportionally great amounts of excrements. Thus, they are condemned to wander around a large area in small groups. This must have been the case with our own species before the advent of agriculture. Because their population size had to remain small, their mitochondrial genomes seldom would have undergone subclonal diversification via ancestral sisters. Therefore, it is in such a population that one might expect to find the parallel persistence of ancient mitochondrial lineages, each ultimately derived from one of the female founders of that population or even of that species. Fortunately, included in the already noted seminal study on human mitochondrial genomes were 15 individuals of the Kung tribe of Africa: hunters and gatherers with an estimated population size of between 5,000 and 10,000 (5). As shown in Fig. 3 Right, which is excerpted from figure 3 of the seminal study (5), 8 ancient stem mitochondrial lineages were embodied in only 15 individuals of the Kung tribe. These stem lineages differed from each other by as much as 2% and as little as 1.8%. It should be recalled that the maximal sequence divergence observed among all of the diverse human beings studied was 2% (5). It may seem strange that this maximal divergence was embodied within a single tribe of hunters and gatherers. It follows that these eight ancient stem lineages must have descended from eight of the female founders (Eves) of our own species. To be sure, there were interferences by sisters even among Kungs. One stem lineage shown in Fig. 3 Right (b) was shared by two individuals, Nos. 9 and 10, whereas the lineage of stem d split into two sublineages, that of stem f split into three, and one sublineage of stem f was shared among four individuals: Nos. 1, 2, 3, and 4. These, however, were clearly much more recent events.
Figure 3.
Two excerpts from figure 3 of the seminal work by Wilson and coworkers (5). (Left) The mitochondrial sequence cluster of three derived from one African American, one Nigerian, and one Asian. (Right) Mitochondrial dendrogram of 15 individuals of the Kung tribe of hunters and gatherers.
The Fourth Rule: Two or More Individuals Not Apparently Related May Share an Identical or Nearly Identical Mitochondrial Genome
When the ancestries of individuals A and B depicted in Fig. 1 are viewed separately, the one ancestor per generation rule of mitochondrial inheritance is illustrated. Viewed as a whole, on the other hand, Fig. 1 also illustrates the fourth and last rule of mitochondrial inheritance. Individuals A and B are related to each other only to 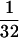 degree, because the two shared one great-, great-, great-grandmother who mated with two genetically unrelated males. Nevertheless, because this great-, great-, great-grandmother was from the maternal half in the ancestry of both A and B, she managed to transmit the identical mitochondrial genome to both individuals. Needless to say, the same would have happened if two maternal half-sisters would have taken the place of this great-, great-, great-grandmother. Within the time span of five generations (not more than 100 years), no sequence diversification would have mired the mitochondrial identity between individuals A and B.
degree, because the two shared one great-, great-, great-grandmother who mated with two genetically unrelated males. Nevertheless, because this great-, great-, great-grandmother was from the maternal half in the ancestry of both A and B, she managed to transmit the identical mitochondrial genome to both individuals. Needless to say, the same would have happened if two maternal half-sisters would have taken the place of this great-, great-, great-grandmother. Within the time span of five generations (not more than 100 years), no sequence diversification would have mired the mitochondrial identity between individuals A and B.
Also seen in figure 3 of the seminal work by Wilson and coworkers (5) was a cluster of three individuals whose mitochondrial sequences differed from each other by only two base substitutions each (0.25% differences). This portion was excerpted in Fig. 3 Left. What is striking is that this cluster is made of one African American, one Nigerian, and one Asian. Clearly, the situation similar to the one depicted in Fig. 1 has actually happened in a more remote past. Of the two ancestral sisters, one remained in Africa to become an ancestor of an African American and a Nigerian, while the other took part in a migration to the Near East in a small band, thereby becoming an ancestor of an Asian.
References
- 1.Ohno S. Proc Natl Acad Sci USA. 1996;93:15276–15278. doi: 10.1073/pnas.93.26.15276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Margolis L. Origin of Eukaryotic Cells. New Haven, CT: Yale Univ. Press; 1970. [Google Scholar]
- 3.John P. Reference. 1987;90:140–150. [Google Scholar]
- 4.Kaneda H, Hayashi J, Takahama S, Taya C, Fischer-Lindahl K, Yonekawa H. Proc Natl Acad Sci USA. 1995;92:4542–4546. doi: 10.1073/pnas.92.10.4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vigilant L, Pennington R, Harpending H, Kocher T D, Wilson A C. Proc Natl Acad Sci USA. 1989;86:9350–9354. doi: 10.1073/pnas.86.23.9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fumihito A, Miyake T, Takada M, Shingu R, Endo T, Gojobori T, Kondo N, Ohno S. Proc Natl Acad Sci USA. 1996;93:6792–6795. doi: 10.1073/pnas.93.13.6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horai S, Hayasaka K, Kondo R, Tsugane K, Takahata N. SMBE 3 Abstracts. Third International Meeting of the Society of Molecular Biology and Evolution; 1995. p. 18. (abstr.). [Google Scholar]