Abstract
Eukaryotic transcriptional activators may function by stimulating formation of RNA polymerase II preinitiation complexes at the core promoter of genes. In this case, their mode of action will intrinsically depend on how these complexes assemble on promoters in living cells, an issue that remains largely unexplored. Here we show that in yeast the basal transcription machinery is brought to the promoter in the form of at least two subcomplexes, TFIID and a complex comprising TFIIB and other essential components. Individual recruitment of either complex by artificial contact with a transcriptionally inactive, sequence-specific DNA-binding protein suffices to trigger transcriptional activation from a wild-type core promoter bearing the appropriate binding site. In contrast, activation from a promoter containing a weakened TATA element is only observed upon recruitment of TFIID. Tethering TFIIB on that promoter remains without effect, but the simultaneous recruitment of both components leads to strong synergistic activation. These findings suggest a simple mechanism whereby two activators that contact distinct subcomplexes of the basal machinery may stimulate transcription synergistically and differentially depending on the nature of the promoter.
Keywords: RNA polymerase II, gene activation, in vivo
A major goal in the field of eukaryotic gene expression is to understand how transcriptional activator proteins stimulate the rate at which RNA polymerase II (pol II)-dependent protein-encoding genes are transcribed. Activators typically contain two physically and functionally separable domains: a DNA-binding domain that recognizes specific regulatory elements in the enhancer of target genes, and an activation domain that stimulates transcription initiated at the adjacent promoter (1). The promoter specificity of activators is conferred both by the specificity of the DNA-binding domain and by the activation domain that can display distinct promoter-selective activation properties (2–5). Efficient transcription of most genes requires a combination of multiple activators that act in concert to stimulate transcription synergistically. The ability of activators to function in a promoter-selective and synergistic manner constitutes the major basis for the enormous diversity of eukaryotic gene expression (6).
Accurate initiation of transcription requires assembly at the promoter of a large preinitiation complex comprising RNA pol II, a collection of general transcription factors (GTFs) including TFIID and TFIIB, and a number of additional polypeptides (7). Activators are believed to function primarily by facilitating formation of functional preinitiation complexes through interactions with one or more components of the basal machinery (8–10). These interactions are thought to recruit, stabilize, and/or induce conformational changes in the preinitiation complex (11, 12). Recent experiments in yeast have shown that artificially recruiting diverse components of the basal machinery to the promoter suffices to stimulate transcription, indicating that conformational changes are not a prerequisite for activation in vivo (13–17). These findings, together with previous work showing that activator function has very limited amino acid sequence requirement (18, 19), are compatible with many natural activators working, at least in part, by recruitment of the basal machinery.
The ability of activators to function in a promoter-selective and synergistic manner will largely depend on how functional transcription complexes assemble on promoters in living cells, an issue that remains uncertain. Earlier in vitro studies led to the dogma that assembly occurs in multiple discrete steps that would each provide a point for regulation. The stepwise model has recently been called into question by the biochemical isolation from both yeast and mammalian cell nuclei of large complexes containing some (20–22), or all (23), of the GTFs and other proteins together with pol II, the so-called holoenzyme. This finding raises the possibility that active transcription complexes may be formed on promoters by recruitment of a single preassembled complex rather than by stepwise recruitment of individual components (24–26). However, while a holoenzyme is certainly the molecular entity that initiates transcription at most promoters in vivo, its varied composition together with the fact that ≈80% of RNA pol II and GTFs are found independent of this complex (27) leaves unanswered the issue of how the entire machinery assembles at the promoter in living cells.
We have undertaken a genetic strategy in yeast the Saccharomyces cerevisiae with the aim of identifying the steps in preinitiation complex formation that may be subject to regulation by activators. Our approach is based on the premise that association of a basal component to a promoter is limiting in preinitiation complex assembly if facilitation of this event by artificial recruitment leads to increased transcription. Using such a strategy, we show here that in yeast the transcription machinery is brought to the promoter in the form of at least two subcomplexes, TFIID and a complex comprising TFIIB and other essential components. The implications of these findings with respect to core promoter selective response to different activators and synergistic activation are discussed.
MATERIALS AND METHODS
Plasmid Constructions.
All the proteins are encoded by single copy plasmids marked with the URA3, TRP1 (28), or ADE2 (E.G.-C. and M.S., unpublished data) genes, with the following exceptions. In Fig. 1, the indicated proteins were overproduced by introducing the genes into the multicopy vector pYES2 (Invitrogen) under control of the GAL1 promoter and by growing transformants in galactose medium. To avoid interference, TFIIB (Figs. 2 and 4) and TAF90 (Fig. 4) were expressed at low levels from pYES2 by growing the cells in medium containing glucose. Max–RFX (RFX in Figs. 1 and 4) and VP16–RFX have been described (14). The regions encoding TAF60 (29), TAF90 (30), and TFIIB (31) were amplified by PCR from yeast genomic DNA using primers that introduced restriction endonuclease sites to allow direct subcloning. TAF60 and TAF90 were fused in-frame to the carboxyl terminus of Max–RFX to generate RFX–TAF60 and RFX–TAF90. TAF90 was also fused to the carboxyl terminus of VP16–RFX to generate VP16–RFX–TAF90. The Myc dimerization motif (14) was joined carboxyl- or amino-terminal of TFIIB to produce TFIIB–Myc and TFIIB–Myc(Nter), respectively. In Fig. 4, TFIIB was directly linked to the amino terminus of RFX. The G204R, F189R, G189R/F294R, and C48S substitutions were introduced into the TFIIB coding region by PCR using appropriate primers and the naturally occurring ScaI and AseI sites. Unless overexpressed, all the RFX derivatives are under control of the TBP promoter. The TFIIB–Myc variants were expressed either from the native TFIIB promoter in the lacZ and viability assays or from ADH1 and DED1 promoter in the Western blot analyses presented in Figs. 2 and 3, respectively. The TFIIB derivatives presented in Fig. 3 were overexpressed from the ADH1 promoter. Details of plasmid constructions are available upon request.
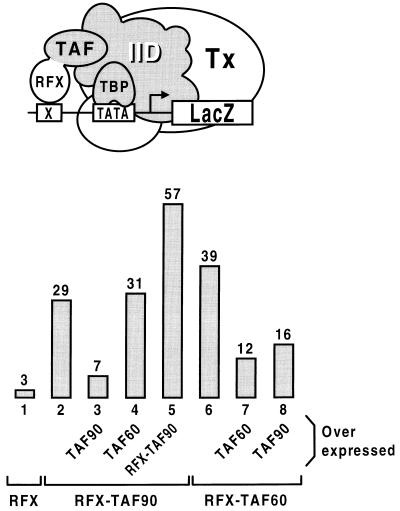
Figure 1.
Recruitment of TFIID to a target promoter by fusing TAF90 or TAF60 to the transcriptionally inactive DNA-binding protein RFX stimulates transcription. The activity of a lacZ reporter gene bearing a single RFX-binding site (X) upstream of the his3 TATA element (14) was assessed in yeast strains expressing the indicated proteins from plasmid DNAs. The proteins were either expressed at low levels from the TBP promoter on a centromeric plasmid, or overproduced by introducing the genes into the multicopy vector pYES2 (Invitrogen) under control of the GAL1 promoter. Transformed yeast cells were grown in selective medium containing galactose and assayed for β-galactosidase activity. In this and the other figures of this paper, values are relative to the level of β-galactosidase activity seen with VP16–RFX under normal conditions: this amount was assigned a value of 100. Tx refers to the remainder of the general factors and pol II.
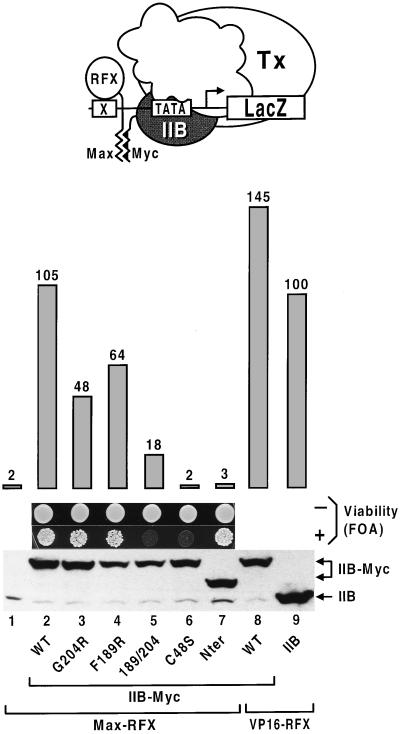
Figure 2.
Transcriptional activation upon recruitment of TFIIB–Myc by Max–RFX. The activity of the RFX-dependent lacZ gene was determined in strains expressing the indicated RFX fusion proteins from the TBP promoter and TFIIB derivatives from the native TFIIB promoter. The parent strain contains a chromosomal TFIIB deletion and expresses low levels of wild-type TFIIB from pYES2 by growing the cells in glucose medium. The ability of the TFIIB–Myc variants to support viability was examined by spotting 104 transformants on plates containing 5-fluoroorotic acid (FOA), a uracil analog that selects against cells carrying pYES2 marked with URA3 and encoding wild-type TFIIB. A protein immunoblot of whole-cell extracts from strains expressing the TFIIB–Myc proteins at higher levels from the ADH1 promoter (lanes 1–8) or TFIIB from its own promoter (lane 9) is shown at the bottom with the positions of TFIIB and TFIIB–Myc indicated. The TFIIB–Myc mutants contain either (lanes 4 and 3) or both (lane 5) F189R, G204R substitutions of residues known from the crystal structure to contact TBP (38), or a single C48S mutation (lane 6) important for interaction with pol II–TFIIF (39–42). Nter designates a hybrid protein carrying the Myc dimerization motif at the amino terminus (lane 7).
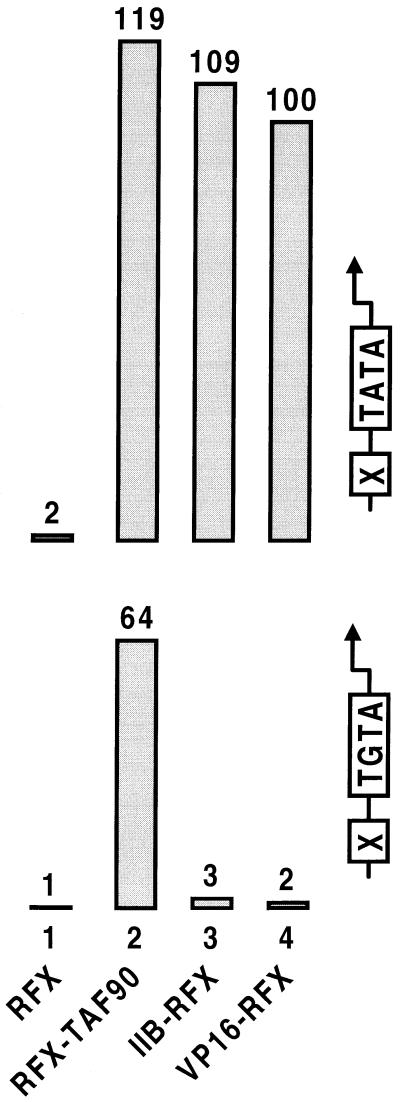
Figure 4.
The nature of the TATA element differentially influences transcriptional activation by recruitment of TFIID or TFIIB. Comparison of transcriptional activation by TAF90 or TFIIB fused covalently to RFX, or by VP16–RFX from wild-type (TATA) and mutant (TGTA) promoters bearing an upstream RFX-binding site (14). The chimeric proteins were expressed at low levels from the TBP promoter on a centromeric plasmid. Because the wild-type proteins interfere with activation mediated by the derivatives fused to RFX (see Figs. 1 and 3), the strains are deleted either for the chromosomal TAF90 gene (lanes 1 and 2), or for the TFIIB gene (lanes 3 and 4), and they express low levels of the corresponding proteins from pYES2 by growing the cells in glucose medium.
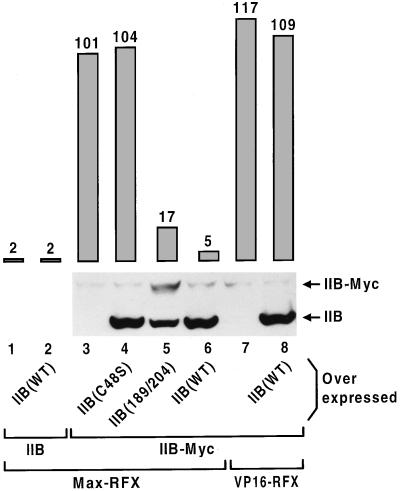
Figure 3.
TFIIB associates with essential proteins before reaching the promoter. Wild-type and mutant forms of TFIIB expressed at high levels from the ADH1 promoter were examined for their effect on transcription activated by TFIIB–Myc and Max–RFX (lanes 4–6), or by VP16–RFX (lane 8). The TFIIB mutants carry either the C48S substitution that compromises interaction of TFIIB with pol II–TFIIF (39–42) (lane 4), or the double amino acid change F189R/G204R on the surface of the protein that contacts TBP (38) (lane 5). The strains containing TFIIB–Myc are deleted for the chromosomal TFIIB gene. The amounts of TFIIB proteins expressed in these cells were determined by the protein immunobloting shown at the bottom.
Yeast Strains and Phenotypic Analyses.
The yeast strains, derived from KY320 (32), carry a lacZ reporter gene integrated at the HIS3 locus with a single RFX-binding site upstream of a consensus or mutated his3 TATA element (14). Where indicated, the chromosomal TFIIB locus was replaced by a version deleted between the ScaI and BamHI sites in the coding region, and the TAF90 locus was disrupted by deleting the coding sequence between the BglII and BamHI sites. The various TFIIB–Myc fusions were tested for their ability to support viability in strains deleted for the chromosomal TFIIB gene and expressing wild-type TFIIB from pYES2 marked with URA3 by spotting 104 cells on plates containing 5-fluoroorotic acid. Transformed yeast cells were grown in selective medium and assayed for β-galactosidase activity as described (33), except that measurements were normalized to the absorbance at 600 nm of the cultures.
Yeast Extracts and Immunoassays.
Whole cell extracts were prepared as described (34), except that the lysis buffer contained 10% glycerol, 20 mM Hepes-KOH (pH 7.9), 350 mM ammonium sulfate, 10 mM EDTA, 1 mM DTT, 0.5 mg/ml BSA, 5 mM benzamidine, 1 mM phenylmethylsulfonyl fluoride, and 10 μg/ml pepstatin, aprotinin, chymostatin, and leupeptin. Proteins were separated on 8% SDS/polyacrylamide gels and electroblotted onto Immobilon-P polyvinylidene difluoride membranes (Millipore). Immunoblotting was performed using the Amersham ECL kit according to the manufacturer, with a 1:250 dilution of purified rabbit anti-yeast TFIIB antiserum and a 1:6,000 dilution of secondary antibody.
RESULTS
Activation by Recruitment of TFIID.
We and others have reported that enhanced recruitment of the TATA box-binding protein (TBP) by artificial interaction with a promoter-bound protein activates transcription in yeast, implying that binding of TBP to at least certain promoters is limiting under physiological conditions (14–16). TBP is the TATA-binding subunit of the larger TFIID complex that also includes TBP-associated factors (TAFs) (9). Although most yeast TAFs are essential for viability, they are apparently dispensable for transcription at most promoters (35, 36), therefore raising the possibility that productive pol II preinitiation complexes lack TAFs in vivo. Genuine components of a functional TFIID complex are expected to stimulate transcription when tethered to the promoter by indirectly recruiting TBP. Fig. 1 shows that TAF90 and TAF60 fused amino terminal to RFX, a human sequence-specific DNA-binding protein with no activation potential in yeast (lane 1 and ref. 14), stimulate transcription of a LacZ reporter gene bearing a single RFX-binding site upstream of the his3 core promoter (lanes 2 and 6). The fusion proteins stimulate transcription to similar levels when tethered to the gal10 core promoter (data not shown). Overexpression of native TAF90 or TAF60 strongly reduces activation by the corresponding hybrid protein (lanes 3 and 7), whereas a higher level of RFX–TAF90 leads to increased transcription, presumably because at lower levels the RFX-binding site is not saturated (lane 5). These effects are most simply interpreted as a competition between the native TAFs and the RFX–TAF fusion proteins for their respective sites in the TFIID complex. In support of this notion is the finding that activation by RFX–TAF90 reaches levels comparable to those observed with VP16–RFX in cells expressing no endogenous TAF90 (see below). We take these results to indicate that the hybrid proteins are part of a functional TFIID complex and, by binding to DNA through their RFX moieties, indirectly recruit TBP to the promoter. Surprisingly, whereas overexpression of TAF60 does not interfere with RFX–TAF90 activation function (lane 4), high levels of TAF90 inhibits activation by RFX–TAF60 (lane 8) while having no effect on viral protein 16 (VP16)-dependent transcription (data not shown). This selective inhibition presumably results from titration of RFX–TAF60 off the TFIID complex by overexpressed TAF90. Thus, TAF60 requires TAF90 to become part of TFIID. Collectively, these results indicate that TAF60 and TAF90 are accessible for interactions with upstream bound factors, and that a TFIID complex comprising TAF60, TAF90, and presumably other TAFs together with TBP, nucleates formation of transcription-competent preinitiation complexes in vivo.
Activation by Recruitment of TFIIB.
We next asked whether transcription can be stimulated by aiding the recruitment of TFIIB to the promoter. This GTF plays a central role within the preinitiation complex by providing a physical link between TFIID and pol II–TFIIF (7). We fused the complementary dimerization domains of the c-Myc oncoprotein and its partner Max (37) to the carboxyl and amino termini of TFIIB and RFX, respectively. TFIIB–Myc functionally substitutes for TFIIB and strongly activates transcription when tethered to a promoter by Max–RFX (Fig. 2, lane 2). Activation occurs only in the presence of Max–RFX (data not shown) and requires TFIIB–Myc to function as a general factor for pol II transcription. Amino acid alterations on the surface of TFIIB that contacts TBP (38), or within the amino-terminal region of the protein essential for interaction with pol II–TFIIF (39–42), compromise the ability of TFIIB–Myc both to sustain cell viability and to respond to Max–RFX while having little effect on protein stability (Fig. 2, lanes 5 and 6). Moreover, a TFIIB variant bearing the Myc dimerization motif at the amino terminus predicted to be close to the start site of transcription, and thus most likely inaccessible for interactions with upstream bound factors (38), is fully competent at supporting cell growth yet unable to promote preinitiation complex formation together with Max–RFX (lane 7). Taken together, these results argue against TFIIB–Myc containing a fortuitous activation domain. Rather, they indicate that the response to Max–RFX of TFIIB–Myc is mediated through its function as a basal factor. We conclude that association of TFIIB with the preinitiation complex is a limiting step in vivo that is strongly stimulated by interaction with a protein bound close to the promoter.
A significant fraction of TFIIB is purified as part of a large multisubunit complex of varied composition, termed the holoenzyme, and containing most (20–22), or all (23), of the GTFs and pol II. To assess whether TFIIB is recruited to the promoter as an isolated subunit or as a component of a preassembled complex, wild-type and mutant forms of TFIIB were tested for their ability to interfere with activation by recruitment of TFIIB–Myc. Fig. 3 shows that high level expression of wild-type TFIIB reduces activation mediated by TFIIB–Myc and Max–RFX to nearly uninduced levels while having no effect on VP16-dependent transcription (lanes 3 versus 6, and 7 versus 8). Activation by TFIIB–Myc is also competed upon overexpression of a TFIIB derivative bearing substitutions in the region that contacts TBP (lane 5), but not with a mutant which binds TBP normally but is defective for interactions with pol II–TFIIF (lane 4). Thus, interference in vivo requires that TFIIB interact with pol II–TFIIF but not with TBP. A similar but less pronounced effect is observed upon expression of the various forms of TFIIB at physiological levels, with a 5- to 10-fold decrease in activation by TFIIB–Myc in cells containing wild-type TFIIB. These findings indicate that TFIIB assembles with other essential components including pol II–TFIIF prior to promoter binding, and that it mediates assembly of productive transcription complexes in vivo mainly in this form.
TFIID and TFIIB Converge to the Promoter Independently.
The observation that TFIIB mutants defective for interaction with TBP maintain the ability to assemble with other essential factors before reaching the promoter does not exclude that TFIID and TFIIB are components of a single complex under normal conditions. To address this issue directly, we compared the effect of tethering either TFIIB or the TAF90 subunit of TFIID to a promoter containing a defective TATA element downstream of the RFX-binding site. If TFIID and TFIIB colocalize in one complex, activation by interaction with either factor should result from recruitment of the same complex and should therefore be equally sensitive to mutations inserted in the TATA element. By contrast, if TFIID and TFIIB were to reach the promoter separately, a direct contact between RFX and TFIID would stabilize weak interactions between TBP and the DNA, and thereby overcome the transcriptional defect caused by mutations in the TATA element. Any recruitment of a TFIIB complex lacking promoter-recognition capabilities would remain without effect because TFIID fails to bind defective TATA elements on its own (43). Fig. 4 shows that TAF90 and TFIIB fused directly to RFX activate transcription comparably to VP16–RFX from a promoter containing a wild-type TATA box and in cells expressing low levels of endogenous TAF90 or TFIIB, respectively. In marked contrast however, transcription from a promoter containing the defective TATA element TGTAAA (and therefore unresponsive to VP16–RFX) is only observed upon recruitment of TFIID (Fig. 4 and ref. 14). Thus, the two factors must reach the promoter as separate entities in vivo.
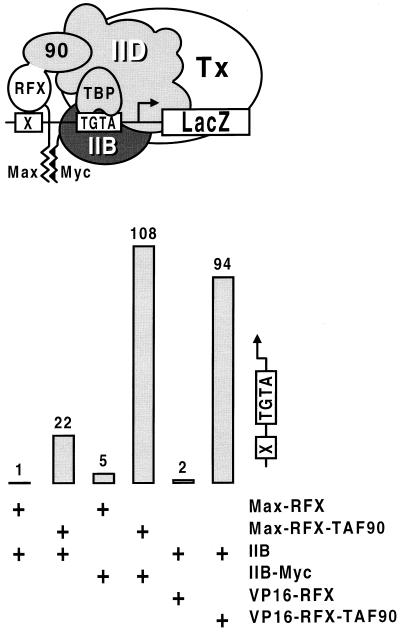
If TFIID and TFIIB converge to the promoter independently, interactions with both factors should activate transcription synergistically, at least on promoters bearing weak TFIID-binding sites. Fig. 5 shows that this is indeed the case. Recruitment of TFIIB–Myc by Max–RFX to a TGTAAA-containing promoter has essentially no effect. But when TFIID is tethered to the promoter together with TFIIB–Myc by fusing covalently TAF90 to Max–RFX, transcription is activated to much higher levels than by recruitment of TFIID alone. Importantly, the effect is mediated by a single RFX hybrid protein that recruits both factors to the promoter through a unique RFX-binding site, therefore excluding the possibility that synergistic activation is accounted for by cooperative DNA-binding of RFX to multiple sites. Consistent with TFIID being mainly preassembled before binding to DNA, tethering both TAF90 and TBP to the mutant promoter has no synergistic effect (data not shown). Thus, while individual contact with either one of the TFIID and TFIIB complexes suffices to trigger strong transcriptional stimulation from a wild-type promoter, efficient activation from a TATA mutant promoter requires interactions with both. Interestingly, tethering TFIID to the TATA mutant promoter also restores its ability to respond to VP16. As both VP16 and the TAF90 subunit of TFIID are covalently fused to the same RFX molecule, VP16 cannot exert its effect by strengthening the interaction between RFX and TFIID. We thus conclude that VP16 can act on steps that follow TFIID binding to DNA (Fig. 5).
Figure 5.
Simultaneous recruitment of TFIID and TFIIB to a TGTAAA-containing promoter activates transcription synergistically. TFIID is tethered to the promoter by fusing covalently TAF90 to Max–RFX, and TFIIB–Myc is recruited by the same Max–RFX–TAF90 hybrid protein through the Max dimerization domain. TAF90 was also fused at the carboxyl terminus of VP16–RFX to assess activity of VP16 upon artificial recruitment of TFIID. The strains used in these experiments carry a deletion of the TFIIB gene, and they express normal levels of endogenous TAF90 which competes with RFX–TAF90 for its site in the TFIID complex, thus explaining the reduced levels of activation mediated by RFX–TAF90 compared with those observed in Fig. 4.
DISCUSSION
Promoter Assembly of an RNA Pol II Transcription Complex.
An increasing body of evidence indicates that eukaryotic activators stimulate pol II transcription in part by recruiting the basal machinery to the promoter of genes. The ability of activators to function in a promoter-selective and synergistic fashion will therefore intrinsically depend on how transcription complexes assemble on promoters in living cells. The analyses reported here provide genetic evidence that in yeast, the basal machinery reaches the promoter in the form of at least two subcomplexes, TFIID and a complex comprising TFIIB and other essential components, that can be recruited independently to the promoter. This is consistent with crystal structures studies of the TBP–TFIIB–DNA and TBP–TFIIA–DNA complexes and site-specific protein–DNA photocrosslinking experiments revealing that the proteins surround promoter DNA to form a cylindrical clamp, thus making it difficult to understand how the entire transcription machinery could be delivered to the promoter in a single step without invoking dramatic conformational changes (38, 44–46). Our results are also in agreement with the most recent biochemical studies showing that the major form of the yeast holoenzyme comprises pol II and several GTFs including TFIIB, but not the TBP subunit of TFIID (47, 48), whereas earlier results had suggested that TBP might be a loosely associated member of the complex (20). Although the TFIIB complex identified in vivo is of unknown composition, it may well correspond to the holoenzyme characterized biochemically. Indeed, we found that entry of TFIIB into the complex requires integrity of the amino-terminal region of the protein, which mediates contact with the RAP30 subunit of TFIIF (41) and is responsible for the recruitment of pol II–TFIIF into the initiation complex in vitro (39, 40). Whether all the other components of the holoenzyme as currently isolated are present in the complex in vivo remains to be determined.
The isolation of distinct forms of holoenzyme that differ in their general factor content, together with the fact that ≈80% of pol II and GTFs are found independent of this complex raise the possibility that a preinitiation complex can assemble through several different pathways in vivo. However, our results indicate that a multiple pathway model is unlikely to apply to TFIIB. First, TFIIB appears to promote formation of productive preinitiation complexes only in association with other essential components (Fig. 3). Interestingly, a TFIIB derivative truncated at the amino terminus and therefore unable to recruit pol II inhibits both basal and VP16-activated transcription in reconstitution experiments by competing with full-length TFIIB for the interaction with TBP bound to DNA (39). By contrast, overexpression of a similar TFIIB mutant in yeast remains without effect, consistent with TFIIB being unable to reach the promoter as an isolated subunit (Fig. 3). Second, the TFIIB complex evidently lacks TFIID as its recruitment on a TATA mutant promoter, whose activity is strictly dependent on the recruitment of TFIID, remains essentially without effect (Fig. 4). Thus, TFIIB appears to mediate assembly of a functional preinitiation complex in vivo only as a component of a preassembled subcomplex that does not include TFIID.
In yeast, TAFs appear to be more loosely associated with TBP than in higher eukaryotes (30, 49), thus raising the possibility that TFIID assembles on the promoter. However, several observations suggest that TBP associates with other proteins important for transcription of at least some essential pol II genes prior to promoter binding. This was first inferred from the finding that dominant-negative mutants of TBP had all lost specifically the ability to bind DNA (50). More recent studies demonstrated that depletion of single yeast TAFs resulted in the selective disappearance of several other yeast TAFs and of yeast TBP, consistent with these proteins existing as a multisubunit complex off the DNA (35). Our results also support the notion that TFIID rather than TBP is the TATA-binding entity in yeast. First, we provide genetic evidence that at least some subunits of TFIID (e.g., TAF60 and TAF90) interact in the absence of DNA (Fig. 1). Second, we find that covalent interactions with both TAF90 and TBP do not activate transcription above the level achieved by contacting either one of these proteins individually, consistent with TFIID being mainly preassembled before reaching the promoter (data not shown, and see below).
Promoter-Selective Activation.
That the transcription machinery is brought to the promoter in the form of at least two subcomplexes, TFIID and a TFIIB-containing complex, has important implications for transcriptional activation. We found that on a wild-type promoter (presumably bearing high affinity binding sites for both components), contact between either one of these components individually and an upstream-bound protein suffices to trigger strong transcriptional activation. As both components are essential for pol II transcription, the one complex tethered to DNA must efficiently recruit the other by binding cooperatively with it at the promoter. By contrast, activation from a promoter bearing a mutated TATA element is only observed upon recruitment of TFIID. Tethering TFIIB on that promoter remains without effect, although recruitment of both components leads to strong synergistic activation (Fig. 4).
Thus, even very strong contact with TFIIB or any other component of the TFIIB complex (by fusion to a heterologous DNA-binding protein or by multiple activator contacts) does not suffice to trigger gene activation at a TATA mutant promoter; a contact with TFIID is also necessary in this case. Were there only one preassembled complex, such a promoter-selective response would not occur. One may speculate that another class of promoter mutants might be identified at which recruitment of TFIID will give very weak activation unless the TFIIB complex is also contacted. In vivo footprinting at the yeast CYC1 promoter revealed protection over the TATA region in the absence of an activator, suggesting that an event distinct from TFIID binding may be limiting at certain promoters (51). Perhaps recruitment of the holoenzyme is required at such promoters for formation of an active transcription complex (13). Interestingly, efficient transcription from a chimeric promoter bearing both types of mutations would strictly depend on contacts with both components.
It is important to stress that, although our experiments involve artificial recruitment of basal factors and the use of mutant promoters, they are likely to be physiologically relevant. Indeed, interactions between natural activators and their targets are likely to be of much lower affinities (52), so that even subtle differences amongst different promoters would nevertheless profoundly influence their response to various activators. Consistent with this view, several examples have already been documented where basal promoter elements serve as selective determinants of activators function (2–5, 53, 54).
Activation by Natural Activators.
In considering the mode of action of natural activators, it is important to keep in mind that any mutation compromising promoter association of one subcomplex should have a stronger effect on activators that function by recruiting the other(s). For example, mutating the TATA element had a much more dramatic effect on activation by recruitment of the TFIIB complex than on activation by recruitment of TFIID (Fig. 4). In this respect, it is striking that the prevalent class of yeast TBP mutants defective in activated transcription of most, but not all genes are selectively impaired in DNA binding, despite the fact that in one reported case the mutants were isolated from libraries biased against the DNA-binding surface of the molecule (55, 56). These mutations in TBP compromise the same TFIID–DNA interaction that is altered by substitutions in the TATA element shown here to impair activation by recruitment of the TFIIB complex more than activation by recruitment of TFIID. It is therefore tempting to speculate that the rate-limiting step created by mutations in the DNA-binding domain of TBP is not affected by the majority of natural activators in the assembly process. We would propose that many activators function in yeast by mainly stimulating a step distinct from TFIID binding to the promoter. Such a scenario would be compatible with yeast TAFs not being generally required for transcription activation (35, 36). It would also be consistent with our observation that VP16 is active on a TATA mutant promoter only if TFIID is simultaneously tethered to DNA.
That TFIID and the remainder of the transcription machinery can also be recruited to the promoter independently in higher eukaryotes is supported by several observations. For example, overexpression of TBP was found to have opposite effects on the activity of different activation domains. Thus, one class of activation domains (exemplified by Sp1) exhibits reduced activity in cells expressing high levels of TBP, whereas another class which includes VP16 becomes more potent under these conditions (57–59). One simple interpretation is that members of the former class function mainly by recruiting TFIID through contacts with TAFs. Inhibition would result as a consequence of overexpressed TBP leading to formation of incomplete TFIID complexes. Activation domains of the second class would act by recruiting a holoenzyme lacking TBP, and would have improved function when TBP concentrations are elevated because the binding of TBP becomes the limiting event for activation in this case. As predicted from our results, members of the second class function poorly on promoters bearing weak TFIID binding sites, and require activators of the first class for full activity (5, 59). Thus, the existence of two or more subcomplexes allows the combinatorial action of multiple activators to stimulate transcription differentially depending on the identity of the promoter sequence and on which subcomplexes of the machinery each individual activator contacts, thereby providing an important mechanism that contributes to the enormous diversity of gene expression.
Acknowledgments
We are most grateful to R. A. Young and M. Hampsey for providing us with anti-TFIIB antisera. We also thank F. Shu and S. K. Burley for advice regarding the TFIIB mutagenesis, and D. Scherly for oligonucleotide synthesis. We thank H. Geiselmann and M. Ptashne for valuable discussions, and M. Cockell, S. Clarkson, and D. Kolakofsky for critical reading of the manuscript. This work was supported by the Swiss National Science Foundation.
ABBREVIATIONS
- pol II
polymerase II
- TBP
TATA box-binding protein
- TAF
TBP-associated factors
- TFIID
-B, -F, transcription factor IID, -B, -F
- GTF
general transcription factor
- VP16
viral protein 16
References
- 1.Ptashne M. Nature (London) 1988;335:683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- 2.Harbury P A, Struhl K. Mol Cell Biol. 1989;9:5298–5304. doi: 10.1128/mcb.9.12.5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wefald F C, Devlin B H, Williams R S. Nature (London) 1990;344:260–262. doi: 10.1038/344260a0. [DOI] [PubMed] [Google Scholar]
- 4.Das G, Hinkley C S, Herr W. Nature (London) 1995;374:657–660. doi: 10.1038/374657a0. [DOI] [PubMed] [Google Scholar]
- 5.Emami K H, Navarre W W, Smale S T. Mol Cell Biol. 1995;15:5906–5916. doi: 10.1128/mcb.15.11.5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Struhl K. Neuron. 1991;7:177–181. doi: 10.1016/0896-6273(91)90256-y. [DOI] [PubMed] [Google Scholar]
- 7.Orphanides G, Lagrange T, Reinberg D. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 8.Kingston R E, Green M R. Curr Biol. 1994;4:325–332. doi: 10.1016/s0960-9822(00)00071-3. [DOI] [PubMed] [Google Scholar]
- 9.Tjian R, Maniatis T. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 10.Ranish J A, Hahn S. Curr Opin Genet Dev. 1996;6:151–158. doi: 10.1016/s0959-437x(96)80044-x. [DOI] [PubMed] [Google Scholar]
- 11.Roberts S G, Green M R. Nature (London) 1994;371:717–720. doi: 10.1038/371717a0. [DOI] [PubMed] [Google Scholar]
- 12.Chi T, Carey M. Genes Dev. 1996;10:2540–2550. doi: 10.1101/gad.10.20.2540. [DOI] [PubMed] [Google Scholar]
- 13.Barberis A, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- 14.Klages N, Strubin M. Nature (London) 1995;374:822–823. doi: 10.1038/374822a0. [DOI] [PubMed] [Google Scholar]
- 15.Chatterjee S, Struhl K. Nature (London) 1995;374:820–822. doi: 10.1038/374820a0. [DOI] [PubMed] [Google Scholar]
- 16.Xiao H, Friesen J D, Lis J T. Mol Cell Biol. 1995;15:5757–5761. doi: 10.1128/mcb.15.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Apone L M, Virbasius C M, Reese J C, Green M R. Genes Dev. 1996;10:2368–2380. doi: 10.1101/gad.10.18.2368. [DOI] [PubMed] [Google Scholar]
- 18.Hahn S. Cell. 1993;72:481–483. doi: 10.1016/0092-8674(93)90064-w. [DOI] [PubMed] [Google Scholar]
- 19.Triezenberg S J. Curr Opin Genet Dev. 1995;5:190–196. doi: 10.1016/0959-437x(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 20.Thompson C M, Koleske A J, Chao D M, Young R A. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 21.Kim Y J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 22.Koleske A J, Young R A. Nature (London) 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 23.Ossipow V, Tassan J P, Nigg E A, Schibler U. Cell. 1995;83:137–146. doi: 10.1016/0092-8674(95)90242-2. [DOI] [PubMed] [Google Scholar]
- 24.Emili A, Ingles C J. Curr Opin Genet Dev. 1995;5:204–209. doi: 10.1016/0959-437x(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 25.Carey M F. Curr Biol. 1995;5:1003–1005. doi: 10.1016/s0960-9822(95)00201-6. [DOI] [PubMed] [Google Scholar]
- 26.Halle J P, Meisterernst M. Trends Genet. 1996;12:161–163. doi: 10.1016/0168-9525(96)30035-8. [DOI] [PubMed] [Google Scholar]
- 27.Koleske A J, Young R A. Trends Biochem Sci. 1995;20:113–116. doi: 10.1016/s0968-0004(00)88977-x. [DOI] [PubMed] [Google Scholar]
- 28.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poon D, Bai Y, Campbell A M, Bjorklund S, Kim Y J, Zhou S, Kornberg R D, Weil P A. Proc Natl Acad Sci USA. 1995;92:8224–8228. doi: 10.1073/pnas.92.18.8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reese J C, Apone L, Walker S S, Griffin L A, Green M R. Nature (London) 1994;371:523–527. doi: 10.1038/371523a0. [DOI] [PubMed] [Google Scholar]
- 31.Pinto I, Ware D E, Hampsey M. Cell. 1992;68:977–988. doi: 10.1016/0092-8674(92)90040-j. [DOI] [PubMed] [Google Scholar]
- 32.Chen W, Struhl K. Proc Natl Acad Sci USA. 1988;85:2691–2695. doi: 10.1073/pnas.85.8.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harshman K D, Moye-Rowley W S, Parker C S. Cell. 1988;53:321–330. doi: 10.1016/0092-8674(88)90393-5. [DOI] [PubMed] [Google Scholar]
- 34.Kolodziej P A, Young R A. Methods Enzymol. 1991;194:508–519. doi: 10.1016/0076-6879(91)94038-e. [DOI] [PubMed] [Google Scholar]
- 35.Walker S S, Reese J C, Apone L M, Green M R. Nature (London) 1996;383:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 36.Moqtaderi Z, Bai Y, Poon D, Weil P A, Struhl K. Nature (London) 1996;383:188–191. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- 37.Amati B, Dalton S, Brooks M W, Littlewood T D, Evan G I, Land H. Nature (London) 1992;359:423–426. doi: 10.1038/359423a0. [DOI] [PubMed] [Google Scholar]
- 38.Nikolov D B, Chen H, Halay E D, Usheva A A, Hisatake K, Lee D K, Roeder R G, Burley S K. Nature (London) 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 39.Barberis A, Muller C W, Harrison S C, Ptashne M. Proc Natl Acad Sci USA. 1993;90:5628–5632. doi: 10.1073/pnas.90.12.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buratowski S, Zhou H. Proc Natl Acad Sci USA. 1993;90:5633–5637. doi: 10.1073/pnas.90.12.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ha I, Roberts S, Maldonado E, Sun X, Kim L U, Green M, Reinberg D. Genes Dev. 1993;7:1021–1032. doi: 10.1101/gad.7.6.1021. [DOI] [PubMed] [Google Scholar]
- 42.Sun Z W, Hampsey M. Proc Natl Acad Sci USA. 1995;92:3127–3131. doi: 10.1073/pnas.92.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wobbe C R, Struhl K. Mol Cell Biol. 1990;10:3859–3867. doi: 10.1128/mcb.10.8.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geiger J H, Hahn S, Lee S, Sigler P B. Science. 1996;272:830–836. doi: 10.1126/science.272.5263.830. [DOI] [PubMed] [Google Scholar]
- 45.Tan S, Hunziker Y, Sargent D F, Richmond T J. Nature (London) 1996;381:127–151. doi: 10.1038/381127a0. [DOI] [PubMed] [Google Scholar]
- 46.Lagrange T, Kim T K, Orphanides G, Ebright Y W, Ebright R H, Reinberg D. Proc Natl Acad Sci USA. 1996;93:10620–10625. doi: 10.1073/pnas.93.20.10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chao D M, Gadbois E L, Murray P J, Anderson S F, Sonu M S, Parvin J D, Young R A. Nature (London) 1996;380:82–85. doi: 10.1038/380082a0. [DOI] [PubMed] [Google Scholar]
- 48.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 49.Poon D, Weil P A. J Biol Chem. 1993;268:15325–15328. [PubMed] [Google Scholar]
- 50.Reddy P, Hahn S. Cell. 1991;65:349–357. doi: 10.1016/0092-8674(91)90168-x. [DOI] [PubMed] [Google Scholar]
- 51.Chen J, Ding M, Pederson D S. Proc Natl Acad Sci USA. 1994;91:11909–11913. doi: 10.1073/pnas.91.25.11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu Y, Reece R J, Ptashne M. EMBO J. 1996;15:3951–3963. [PMC free article] [PubMed] [Google Scholar]
- 53.Simon M C, Fisch T M, Benecke B J, Nevins J R, Heintz N. Cell. 1988;52:723–729. doi: 10.1016/0092-8674(88)90410-2. [DOI] [PubMed] [Google Scholar]
- 54.Homa F L, Glorioso J C, Levine M. Genes Dev. 1988;2:40–53. doi: 10.1101/gad.2.1.40. [DOI] [PubMed] [Google Scholar]
- 55.Arndt K M, Ricupero Hovasse S, Winston F. EMBO J. 1995;14:1490–1497. doi: 10.1002/j.1460-2075.1995.tb07135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee M, Struhl K. Mol Cell Biol. 1995;15:5461–5469. doi: 10.1128/mcb.15.10.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keaveney M, Berkenstam A, Feigenbutz M, Vriend G, Stunnenberg H G. Nature (London) 1993;365:562–566. doi: 10.1038/365562a0. [DOI] [PubMed] [Google Scholar]
- 58.Sadovsky Y, Webb P, Lopez G, Baxter J D, Fitzpatrick P M, Gizang Ginsberg E, Cavailles V, Parker M G, Kushner P J. Mol Cell Biol. 1995;15:1554–1563. doi: 10.1128/mcb.15.3.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Steger G, Ham J, Lefebvre O, Yaniv M. EMBO J. 1995;14:329–340. doi: 10.1002/j.1460-2075.1995.tb07007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]