Abstract
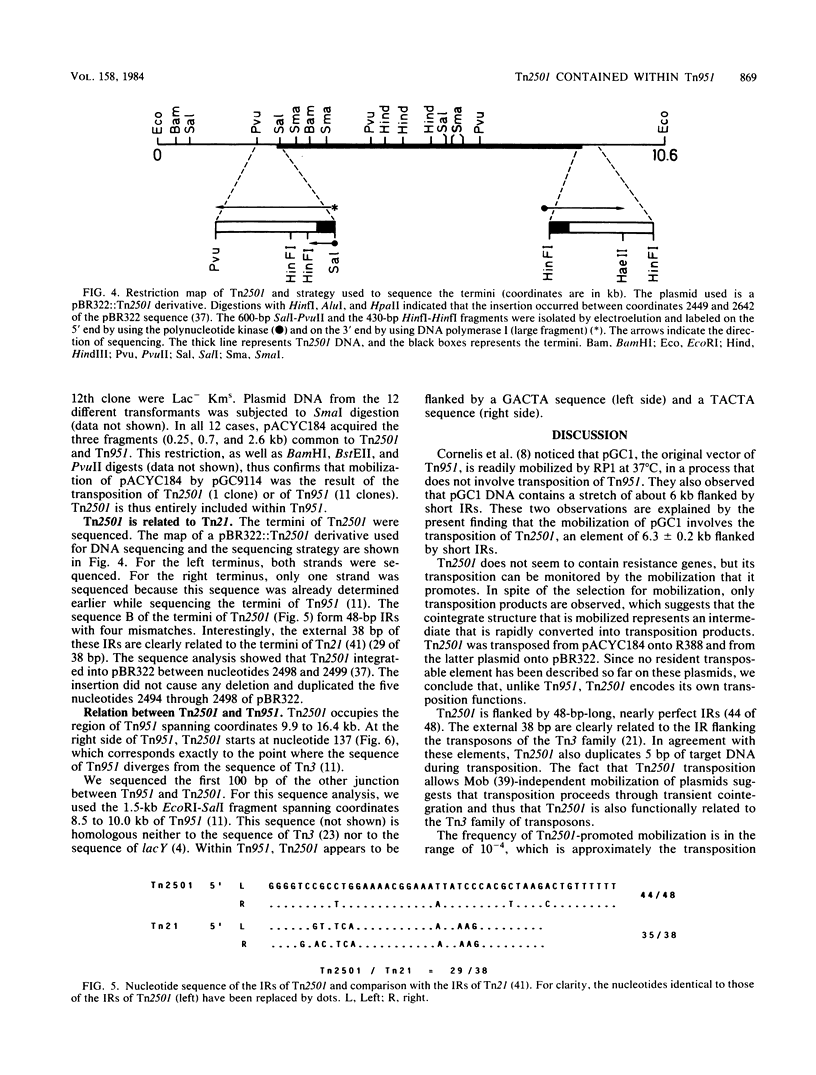
The DNA sequence spanning coordinates 9.9 to 16.4 kilobases of the lactose transposon Tn951 ( Cornelis et al., Mol. Gen. Genet. 160:215-224, 1978) constitutes a transposable element by itself. Unlike Tn951 ( Cornelis et al., Mol. Gen. Genet. 184:241-248, 1981), this element, called Tn2501 , transposes in the absence of any other transposon. Transposition of Tn2501 proceeds through transient cointegration and duplicates 5 base pairs of host DNA. Tn2501 is flanked by nearly perfect inverted repeats (44 of 48), related to the inverted repeats of Tn21 ( Zheng et al., Nucleic Acids Res. 9:6265-6278, 1982). Unlike Tn21 , Tn2501 does not confer mercury resistance.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennett P. M., Grinsted J., Choi C. L., Richmond M. H. Characterisation of Tn501, a transposon determining resistance to mercuric ions. Mol Gen Genet. 1978 Feb 7;159(1):101–106. doi: 10.1007/BF00401753. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Büchel D. E., Gronenborn B., Müller-Hill B. Sequence of the lactose permease gene. Nature. 1980 Feb 7;283(5747):541–545. doi: 10.1038/283541a0. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis G., Bennett P. M., Grinsted J. Properties of pGC1, a lac plasmid orginating in Yersinia enterocolitica 842. J Bacteriol. 1976 Sep;127(3):1058–1062. doi: 10.1128/jb.127.3.1058-1062.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis G., Ghosal D., Saedler H. Multiple integration sites for the lactose transposon Tn 951 on plasmid RP 1 and establishment of a coordinate system for Tn 951. Mol Gen Genet. 1979 Jan 5;168(1):61–67. doi: 10.1007/BF00267934. [DOI] [PubMed] [Google Scholar]
- Cornelis G., Ghosal D., Saedler H. Tn951: a new transposon carrying a lactose operon. Mol Gen Genet. 1978 Apr 6;160(2):215–224. doi: 10.1007/BF00267484. [DOI] [PubMed] [Google Scholar]
- Cornelis G., Saedler H. Deletions and an inversion induced by a resident IS1 of the lactose transposon Tn951. Mol Gen Genet. 1980;178(2):367–374. doi: 10.1007/BF00270486. [DOI] [PubMed] [Google Scholar]
- Cornelis G. Sequence relationships between plasmids carrying genes for lactose utilization. J Gen Microbiol. 1981 May;124(1):91–97. doi: 10.1099/00221287-124-1-91. [DOI] [PubMed] [Google Scholar]
- Cornelis G., Sommer H., Saedler H. Transposon Tn951 (TnLac) is defective and related to Tn3. Mol Gen Genet. 1981;184(2):241–248. doi: 10.1007/BF00272911. [DOI] [PubMed] [Google Scholar]
- Datta N., Hedges R. W. Trimethoprim resistance conferred by W plasmids in Enterobacteriaceae. J Gen Microbiol. 1972 Sep;72(2):349–355. doi: 10.1099/00221287-72-2-349. [DOI] [PubMed] [Google Scholar]
- Depicker A., De Block M., Inzé D., Van Montagu M., Schell J. IS-like element IS8 in RP4 plasmid and its involvement in cointegration. Gene. 1980 Sep;10(4):329–338. doi: 10.1016/0378-1119(80)90153-5. [DOI] [PubMed] [Google Scholar]
- Diver W. P., Grinsted J., Fritzinger D. C., Brown N. L., Altenbuchner J., Rogowsky P., Schmitt R. DNA sequences of and complementation by the tnpR genes of Tn21, Tn501 and Tn1721. Mol Gen Genet. 1983;191(2):189–193. doi: 10.1007/BF00334812. [DOI] [PubMed] [Google Scholar]
- Grindley N. D., Lauth M. R., Wells R. G., Wityk R. J., Salvo J. J., Reed R. R. Transposon-mediated site-specific recombination: identification of three binding sites for resolvase at the res sites of gamma delta and Tn3. Cell. 1982 Aug;30(1):19–27. doi: 10.1016/0092-8674(82)90007-1. [DOI] [PubMed] [Google Scholar]
- Grinsted J., Saunders J. R., Ingram L. C., Sykes R. B., Richmond M. H. Properties of a R factor which originated in Pseudomonas aeruginosa 1822. J Bacteriol. 1972 May;110(2):529–537. doi: 10.1128/jb.110.2.529-537.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinsted J., de la Cruz F., Altenbuchner J., Schmitt R. Complementation of transposition of tnpA mutants of Tn3, Tn21, Tn501, and Tn1721. Plasmid. 1982 Nov;8(3):276–286. doi: 10.1016/0147-619x(82)90065-8. [DOI] [PubMed] [Google Scholar]
- Guyer M. S. The gamma delta sequence of F is an insertion sequence. J Mol Biol. 1978 Dec 15;126(3):347–365. doi: 10.1016/0022-2836(78)90045-1. [DOI] [PubMed] [Google Scholar]
- Hedges R. W., Jacob A. E. Transposition of ampicillin resistance from RP4 to other replicons. Mol Gen Genet. 1974;132(1):31–40. doi: 10.1007/BF00268228. [DOI] [PubMed] [Google Scholar]
- Heffron F., Bedinger P., Champoux J. J., Falkow S. Deletions affecting the transposition of an antibiotic resistance gene. Proc Natl Acad Sci U S A. 1977 Feb;74(2):702–706. doi: 10.1073/pnas.74.2.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron F., McCarthy B. J., Ohtsubo H., Ohtsubo E. DNA sequence analysis of the transposon Tn3: three genes and three sites involved in transposition of Tn3. Cell. 1979 Dec;18(4):1153–1163. doi: 10.1016/0092-8674(79)90228-9. [DOI] [PubMed] [Google Scholar]
- Kado C. I., Liu S. T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981 Mar;145(3):1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecko D. J., Brevet J., Cohen S. N. Involvement of multiple translocating DNA segments and recombinational hotspots in the structural evolution of bacterial plasmids. J Mol Biol. 1976 Dec;108(2):333–360. doi: 10.1016/s0022-2836(76)80124-6. [DOI] [PubMed] [Google Scholar]
- LEDERBERG J. Prevalence of Escherichia coli strains exhibiting genetic recombination. Science. 1951 Jul 20;114(2951):68–69. doi: 10.1126/science.114.2951.68. [DOI] [PubMed] [Google Scholar]
- Lederberg E. M. Plasmid reference center registry of transposon (Tn) allocations through July 1981. Gene. 1981 Dec;16(1-3):59–61. doi: 10.1016/0378-1119(81)90060-3. [DOI] [PubMed] [Google Scholar]
- Low B. Rapid mapping of conditional and auxotrophic mutations in Escherichia coli K-12. J Bacteriol. 1973 Feb;113(2):798–812. doi: 10.1128/jb.113.2.798-812.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McCormick M., Wishart W., Ohtsubo H., Heffron F., Ohtsubo E. Plasmid cointegrates and their resolution mediated by transposon Tn3 mutants. Gene. 1981 Nov;15(2-3):103–118. doi: 10.1016/0378-1119(81)90120-7. [DOI] [PubMed] [Google Scholar]
- Novick R. P., Clowes R. C., Cohen S. N., Curtiss R., 3rd, Datta N., Falkow S. Uniform nomenclature for bacterial plasmids: a proposal. Bacteriol Rev. 1976 Mar;40(1):168–189. doi: 10.1128/br.40.1.168-189.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo H., Ohmori H., Ohtsubo E. Nucleotide-sequence analysis of Tn3 (ap): implications for insertion and deletion. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1269–1277. doi: 10.1101/sqb.1979.043.01.144. [DOI] [PubMed] [Google Scholar]
- Schöffl F., Arnold W., Pühler A., Altenbuchner J., Schmitt R. The tetracycline resistance transposons Tn1721 and Tn1771 have three 38-base-pair repeats and generate five-base-pair direct repeats. Mol Gen Genet. 1981;181(1):87–94. doi: 10.1007/BF00339010. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Hsu M. T., Otsubo E., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. I. Structure of F-prime factors. J Mol Biol. 1972 Nov 14;71(2):471–497. doi: 10.1016/0022-2836(72)90363-4. [DOI] [PubMed] [Google Scholar]
- Sherratt D., Arthur A., Burke M. Transposon-specified, site-specific recombination systems. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):275–281. doi: 10.1101/sqb.1981.045.01.040. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Tu C. P., Cohen S. N. Effect of DNA sequences adjacent to the termini of Tn3 on sequential translocation. Mol Gen Genet. 1980;177(4):597–601. doi: 10.1007/BF00272669. [DOI] [PubMed] [Google Scholar]
- Warren G., Sherratt D. Complementation of transfer deficient ColE1 mutants. Mol Gen Genet. 1977 Mar 7;151(2):197–201. doi: 10.1007/BF00338695. [DOI] [PubMed] [Google Scholar]
- Willetts N. S. Recombination and the Escherichia coli K-12 sex factor F. J Bacteriol. 1975 Jan;121(1):36–43. doi: 10.1128/jb.121.1.36-43.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z. X., Chandler M., Hipskind R., Clerget M., Caro L. Dissection of the r-determinant of the plasmid R100.1: the sequence at the extremities of Tn21. Nucleic Acids Res. 1981 Dec 11;9(23):6265–6278. doi: 10.1093/nar/9.23.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz F., Grinsted J. Genetic and molecular characterization of Tn21, a multiple resistance transposon from R100.1. J Bacteriol. 1982 Jul;151(1):222–228. doi: 10.1128/jb.151.1.222-228.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]





