Abstract
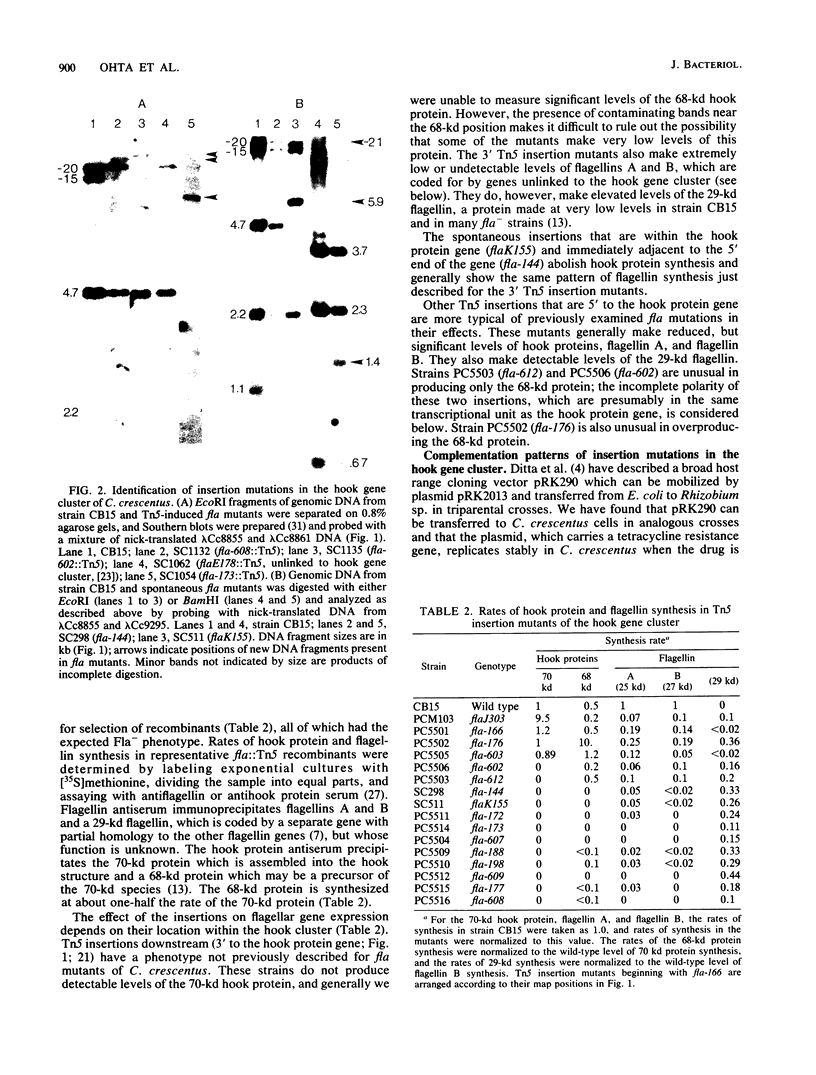
Using the cloned DNA from the hook protein gene region of Caulobacter crescentus ( Ohta et al., Proc. Natl. Acad. Sci. U.S.A. 79:4863-4867, 1982), we have identified and physically mapped 19 Tn5-induced and 2 spontaneous insertion mutations to this region of the chromosome. These nonmotile mutants define a major cluster of fla genes that covers approximately 17 kilobases on the chromosome (hook gene cluster). Complementation analysis of the mutants using DNA fragments from the region subcloned in the broad host range plasmid pRK290 has shown that these fla genes are organized into at least five transcriptional units (I to V). Transcriptional unit II contains at least one gene in addition to the hook protein gene, which makes this the first operon described in C. crescentus. Expression of the hook protein gene and the genetically unlinked flagellin A and B genes by this set of mutants also furnishes additional insights into the hierarchial regulation of flagellar genes. We have found that the spontaneous insertion mutant ( SC511 ) of the hook protein gene ( flaK ) makes no flagellin A or B and that genes downstream from the hook protein gene are required in trans for expression of the hook protein operon and the flagellin A and B genes. Recombination and complementation results thus place flaK , flaJ , flaN , and flaO (R. C. Johnson and B. Ely , J. Bacteriol . 137:627-634, 1979) in the hook gene cluster, identify at least three new genes ( flbD , flbG , and flbF ), and suggest that this cluster may contain several additional, as yet unidentified, fla genes.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berg D. E., Weiss A., Crossland L. Polarity of Tn5 insertion mutations in Escherichia coli. J Bacteriol. 1980 May;142(2):439–446. doi: 10.1128/jb.142.2.439-446.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciampi M. S., Schmid M. B., Roth J. R. Transposon Tn10 provides a promoter for transcription of adjacent sequences. Proc Natl Acad Sci U S A. 1982 Aug;79(16):5016–5020. doi: 10.1073/pnas.79.16.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B., Croft R. H. Transposon mutagenesis in Caulobacter crescentus. J Bacteriol. 1982 Feb;149(2):620–625. doi: 10.1128/jb.149.2.620-625.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill P. R., Agabian N. The nucleotide sequence of the Mr = 28,500 flagellin gene of Caulobacter crescentus. J Biol Chem. 1983 Jun 25;258(12):7395–7401. [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huguenel E. D., Newton A. Localization of surface structures during procaryotic differentiation: role of cell division in Caulobacter crescentus. Differentiation. 1982;21(2):71–78. doi: 10.1111/j.1432-0436.1982.tb01199.x. [DOI] [PubMed] [Google Scholar]
- Iino T. Genetics of structure and function of bacterial flagella. Annu Rev Genet. 1977;11:161–182. doi: 10.1146/annurev.ge.11.120177.001113. [DOI] [PubMed] [Google Scholar]
- Johnson R. C., Ely B. Analysis of nonmotile mutants of the dimorphic bacterium Caulobacter crescentus. J Bacteriol. 1979 Jan;137(1):627–634. doi: 10.1128/jb.137.1.627-634.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Ely B. Isolation of spontaneously derived mutants of Caulobacter crescentus. Genetics. 1977 May;86(1):25–32. doi: 10.1093/genetics/86.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Ferber D. M., Ely B. Synthesis and assembly of flagellar components by Caulobacter crescentus motility mutants. J Bacteriol. 1983 Jun;154(3):1137–1144. doi: 10.1128/jb.154.3.1137-1144.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen R. A., Rothstein S. J., Reznikoff W. S. A restriction enzyme cleavage map of Tn5 and location of a region encoding neomycin resistance. Mol Gen Genet. 1979;177(1):65–72. doi: 10.1007/BF00267254. [DOI] [PubMed] [Google Scholar]
- Katz L., Kingsbury D. T., Helinski D. R. Stimulation by cyclic adenosine monophosphate of plasmid deoxyribonucleic acid replication and catabolite repression of the plasmid deoxyribonucleic acid-protein relaxation complex. J Bacteriol. 1973 May;114(2):577–591. doi: 10.1128/jb.114.2.577-591.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeda Y. Fusions of flagellar operons to lactose genes on a mu lac bacteriophage. J Bacteriol. 1982 Apr;150(1):16–26. doi: 10.1128/jb.150.1.16-26.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milhausen M., Agabian N. Caulobacter flagellin mRNA segregates asymmetrically at cell division. Nature. 1983 Apr 14;302(5909):630–632. doi: 10.1038/302630a0. [DOI] [PubMed] [Google Scholar]
- Milhausen M., Gill P. R., Parker G., Agabian N. Cloning of developmentally regulated flagellin genes from Caulobacter crescentus via immunoprecipitation of polyribosomes. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6847–6851. doi: 10.1073/pnas.79.22.6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton A. Role of transcription in the temporal control of development in Caulobacter crescentus (stalk-rifampin-RNA synthesis-DNA synthesis-motility). Proc Natl Acad Sci U S A. 1972 Feb;69(2):447–451. doi: 10.1073/pnas.69.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta N., Chen L. S., Newton A. Isolation and expression of cloned hook protein gene from Caulobacter crescentus. Proc Natl Acad Sci U S A. 1982 Aug;79(16):4863–4867. doi: 10.1073/pnas.79.16.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POINDEXTER J. S. BIOLOGICAL PROPERTIES AND CLASSIFICATION OF THE CAULOBACTER GROUP. Bacteriol Rev. 1964 Sep;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purucker M., Bryan R., Amemiya K., Ely B., Shapiro L. Isolation of a Caulobacter gene cluster specifying flagellum production by using nonmotile Tn5 insertion mutants. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6797–6801. doi: 10.1073/pnas.79.22.6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Ruvkun G. B., Sundaresan V., Ausubel F. M. Directed transposon Tn5 mutagenesis and complementation analysis of Rhizobium meliloti symbiotic nitrogen fixation genes. Cell. 1982 Jun;29(2):551–559. doi: 10.1016/0092-8674(82)90171-4. [DOI] [PubMed] [Google Scholar]
- Shapiro L. Differentiation in the Caulobacter cell cycle. Annu Rev Microbiol. 1976;30:377–407. doi: 10.1146/annurev.mi.30.100176.002113. [DOI] [PubMed] [Google Scholar]
- Sheffery M., Newton A. Purification and characterization of a polyhook protein from Caulobacter crescentus. J Bacteriol. 1979 May;138(2):575–583. doi: 10.1128/jb.138.2.575-583.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffery M., Newton A. Regulation of periodic protein synthesis in the cell cycle: control of initiation and termination of flagellar gene expression. Cell. 1981 Apr;24(1):49–57. doi: 10.1016/0092-8674(81)90500-6. [DOI] [PubMed] [Google Scholar]
- Silverman M. Building bacterial flagella. Q Rev Biol. 1980 Dec;55(4):395–408. doi: 10.1086/411982. [DOI] [PubMed] [Google Scholar]
- Silverman M., Simon M. I. Bacterial flagella. Annu Rev Microbiol. 1977;31:397–419. doi: 10.1146/annurev.mi.31.100177.002145. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]