Abstract
In the present work, we have asked whether a group of 13 essential genes mapping to the heterochromatin of Drosophila melanogaster chromosome 2 are mutable following transposition of the I factor during I-R hybrid dysgenesis. We found that the frequency of lethal events mapping to chromosome 2 heterochromatin is surprisingly high, despite the low density of genetic functions identified in this region compared with euchromatin. Cytogenetic and molecular analyses indicated that the recovered mutations correspond either to insertions or to rearrangements. Moreover, chromosomes bearing specific heterochromatic lethal mutations were generated by recombination in the heterochromatin. Together, these data indicate that I factors transpose with high frequency into pericentric regions of chromosome 2 and may play a role in the evolution of constitutive heterochromatin.
Transposable elements (TEs) are repetitive mobile DNA sequences identified in a wide variety of organisms, ranging from bacteria to humans. These elements generally constitute 10–20% of the total eukaryotic genome (1). Besides their euchromatic location, TE-homologous sequences are frequently found in the heterochromatin of many species, including humans (2–7). In particular, recent studies have clearly established that in Drosophila melanogaster several clusters of retrotransposon-homologous sequences are present throughout the constitutive heterochromatin of all chromosomes (8).
TE accumulation represents one of the most intriguing aspects of the structure and organization of Drosophila heterochromatin. It has been suggested that heterochromatic regions may contain hotspots for TE insertions (9). Alternatively, the accumulation of TEs into heterochromatin might result from the combined effect of the genetic inertness and the inability of this part of the genome to undergo meiotic recombination (10); according to the latter hypothesis, transposition into constitutive heterochromatin would generally not give rise to mutational events with detrimental effects on fitness. However, D. melanogaster heterochromatin contains several single-copy and repetitive genetic loci (11) which a priori should not escape TE-mediated mutagenesis. Although it is well established that TEs are a major source of spontaneous euchromatic mutations in D. melanogaster, little is known about mutability of heterochromatic genes following transposition. Only a few studies have shown that normal or modified P elements, which mobilize through a DNA intermediate, are able to insert into the heterochromatin and can indeed cause insertional mutations (12–14). In addition, R1 and R2 retroelements insert at specific sites in the heterochromatic 28S rRNA genes of Drosophila (15), thus inactivating transcription of the rDNA unit. These elements belong to the evolutionarily widespread family of non-long terminal repeat (non-LTR) elements which are mobilized by reverse transcription of an RNA intermediate. Representatives of this category also include mammalian L1 sequences and both the Het-A elements and I factors of Drosophila (16–18). The D. melanogaster I factor belongs to the class of long interspersed nuclear elements (LINEs) and actively transposes in the germ line of the female progeny from crosses between reactive (R) females and inducer (I) males. This phenomenon, known as the I-R hybrid dysgenesis, gives rise to reduced fertility, increased frequency of euchromatic mutations, and X chromosome loss (19–21). Inducer strains contain 10–15 euchromatic dispersed copies of the 5.4-kb functional I factor, while several defective copies of I are embedded into the heterochromatin of both inducer and reactive strains (7, 20, 21). However, it remains uncertain whether the I factor can efficiently transpose in the heterochromatin.
To investigate LINE transposition in the heterochromatin and its mutagenic effect on heterochromatic genes, we have studied the mutability of a group of 13 vital loci mapping to the pericentric regions of D. melanogaster chromosome 2, following transposition of the I factor in the I-R hybrid dysgenesis. These genes have been extensively characterized at both the genetic and cytological levels and were found to behave like unique sequences (22–24). It has been shown that at least three of them, light (lt), concertina (cta), and rolled (rl), are indeed single-copy genes (11, 25). Our results show that the heterochromatic genes of chromosome 2 are highly mutable in I-R dysgenesis, and they provide evidence for a causal relationship between I factor transposition and heterochromatic mutability.
MATERIALS AND METHODS
Drosophila Stocks.
Genetic markers, mutations, and special chromosomes used in this work were described by Lindsley and Zimm (26). Genetic crosses were performed at 20°C ± 1°C. Reactive and inducer strains, which lack P elements, were provided by C. Vaury (Institut National de la Santé et de la Recherche Médicale, Clermont-Ferrand, France) and A. Pelisson (Centre National de la Recherche Scientifique, Gif-sur-Yvette, France).
Recovery of I-R Hybrid Dysgenesis-Induced Heterochromatic Lethal Mutations.
Five dysgenic crosses involving the Charolles and cn reactive strains and the Canton-S, Cy/Pm, M-5, and b lt rl cn inducer strains were designed (Table 1). In these crosses, females from a reactive strain were crossed to males from an inducer strain; the F1 dysgenic females were then mated to CyO, S/Sco males. F2 males, heterozygous for the CyO, S balancer and its homolog transmitted from dysgenic females, were individually mated to Df(2R)M-S210/Cy or Df(2L)Sdlt5/Cy females. Df(2R)M-S210 removes the bulk of the right arm heterochromatin of chromosome 2 (2Rh) with its vital genes, while Df(2L)Sdllt5 removes only the distal portion of the left arm heterochromatin of chromosome 2 (2Lh), a region which contains seven vital genes (24). The absence of Cy+ progeny in the offspring of this cross indicates the induction of a lethal mutation on chromosome 2 which is uncovered by the heterochromatic deficiency. Chromosomes that failed to complement Df(2L)Sdlt5 were also tested over Df(2L)C′, which also lacks all the 2Lh genes (22). Chromosomes carrying lethal mutations mapping to heterochromatin were then tested for their ability to complement ethyl methanesulfonate (EMS)-induced alleles representing 13 genetic loci in 2Lh and 2Rh. For each complementation tests 150–200 progeny were counted. Among the chromosomes with 2Lh lethal alleles, IR10 and IR12 lines exhibited low viability and fertility and were subsequently lost. In the control crosses nondysgenic females were obtained by crossing inducer females to reactive males.
Table 1.
Heterochromatic lethals of chromosome 2 induced by I-R dysgenesis
| Cross | Het. | Chr. | Lethals | Frequency | |
|---|---|---|---|---|---|
| 1a | 2R | 1,564 | 11 | 7 × 10−3 | |
| 1b | 2R | 1,003 | 0 | 0 | |
| 2a | 2L | 1,102 | 8 | 7.2 × 10−3 | |
| 2b | 2L | 1,131 | 0 | 0 | |
| 3a | 2R | 1,204 | 5 | 4.1 × 10−3 | |
| 3b | 2R | 896 | 0 | 0 | |
| 4a | 2R | 919 | 2 | 2.2 × 10−3 | |
| 4b | 2R | 381 | 0 | 0 | |
| 5a | 2R | 1,024 | 2 | 1.9 × 10−3 | |
| 5b | 2R | 967 | 1 | 1 × 10−3 | |
| 6a | 2L | 909 | 2 | 2.2 × 10−3 | |
| 6b | 2L | 712 | 0 | 0 | |
| 7a | 2R | 1,142 | 6 | 5.2 × 10−3 | |
| 7b | 2R | 993 | 0 | 0 | |
| 8a | 2L | 746 | 1 | 1.3 × 10−3 | |
| 8b | 2L | 520 | 0 | 0 | |
| Exp. | 2R | 5,853 | 26 | 4.4 × 10−3 | χ2 = 16.24 |
| C. | 2R | 4,240 | 1 | 2.3 × 10−4 | P ≤ 0.001 |
| Exp. | 2L | 2,757 | 11 | 4 × 10−3 | χ2 = 11.17 |
| C. | 2L | 2,363 | 0 | 0 | P ≤ 0.001 |
Crosses 1a and 2a = Ch (R) × C-S (I); 3a = Ch (R) × M-5 (I); 4a = cn (R) × Cy/Pm (I); 5a and 6a = cn (R) × C-S (I); 7a and 8a = Ch (R) × b lt rl cn (I), in which Ch = Charolles; C-S = Canton-S. The reciprocal control crosses are indicated with b. Het., the heterochromatic location of lethal mutations as judged by complementation with 2Lh or 2Rh deletions; Chr., number of chromosomes scored; Lethals, number of lethal chromosomes; Frequency, frequency of lethal chromosomes; Exp., total dysgenic crosses; C., total nondysgenic controls.
To compare the frequency of euchromatic vs. heterochromatic lethals simultaneously induced along chromosome 2, Charolles (R) females were crossed to Canton-S (I) males and the F1 dysgenic females were then mated to CyO, S/Sco males. F2 males, heterozygous for the CyO, S balancer and its homolog transmitted from dysgenic females, were individually crossed to CyO, S/Sco females to establish several lines. Homozygous lethal chromosomes were then tested for their ability to complement EMS-induced alleles representing 13 heterochromatic genes of chromosome 2.
Recovery of Heterochromatic Recombinants.
Dysgenic females were recovered by crossing wild-type Charolles (R) females to b lt rl cn/b lt rl cn (I) males. In the control crosses b lt rl cn/+ + + + nondysgenic females were obtained by crossing b lt rl cn/b lt rl cn (I) females to wild-type Charolles (R) males. The F1 b lt rl cn/+ + + + dysgenic and nondysgenic females were then mated to b lt rl cn/b lt rl cn (I) males; the frequencies of euchromatic and heterochromatic recombinants in the F2 offspring were then calculated. Recombinant lines were established by crossing single b lt + +/b lt rl cn and b lt rl +/b lt rl cn males to Cy, cn/Sco females, and + + rl cn/b lt rl cn males to Sco lt rl/Cy females. The b lt rl cn/+ + + + females were also used to determine the frequency of heterochromatic lethals on chromosome 2 (Table 1: crosses 7a–7b and 8a–8b). The chromosomes with heterochromatic lethal alleles recovered in these experiments were backcrossed to b lt rl cn homozygous flies to establish whether they had been induced on chromosomes that had undergone heterochromatic recombination.
Nucleic Acid Manipulations, DNA Sequencing, PCR Amplification, and Cloning.
DNA and RNA extraction, Southern and Northern hybridization assays, and recombinant DNA techniques were essentially performed according to standard procedures (27). The I factor insertion in the intron 2 of the rl gene was amplified by PCR (30 cycles; 94°C for 1 min, 55°C for 1 min, 72°C for 3 min) using 0.1 μg of genomic DNA from the Canton-S, Charolles, and rlIR1 strains, and the following oligonucleotide primers: rl3 (5′-GCGGATGACACGCTAACA-3′), derived from exon 2 of the rl gene (M95124, nucleotides 415–432), and I5R (5′-TGCTGTTACATCCTCATCTG-3′), derived from the 5′ end of the I factor (M14954, nucleotides 561–542). The 982-bp PCR product obtained from the rlIR1 strain was subcloned into the EcoRV site of the pBC KS (+) vector (Stratagene) and sequenced with the following primers: (i) T3; (ii) T7; (iii) I5R2 (5′-GGATTAGCGGTATTGTTG-3′, M14954 nucleotides 260–243); (iv) rlint2 (5′-CAGGCATACCCCATGAGG-3′), and (v) B290 (5′-TCGAAAGAGTTGTTGTCA-3′, M14954 nucleotides 392–375). The hybridization probes were a 1.4-kb cDNA HindIII restriction fragment containing the complete coding region of the rl gene (28), a 1.5-kb SalI–HindIII fragment containing the 5′ end of the I factor (20), and a 530-bp HindIII–DraI fragment containing the 5′ untranslated region and the first two exons of the rl cDNA (W. H. Biggs & K. Zavitz, personal communication).
RESULTS
Heterochromatic Mutations Induced by I-R Hybrid Dysgenesis.
Our aim was to investigate whether activation of I factor transposition resulted in genetic instability of the heterochromatin. Five different dysgenic crosses were set up, and 37 lethal chromosomes 2 were recovered (Table 1). Of these, 26 of 5,853 chromosomes (4.4 × 10−3) failed to complement Df(2R)M-S210, thus mapping the lethal alleles to 2Rh, while the remaining 11, recovered from a total of 2,757 chromosomes (4 × 10−3), failed to complement Df(2L)Sdlt5 and mapped to 2Lh. In nondysgenic controls, only one lethal mapping to 2Rh was recovered among 4,240 chromosomes, while no lethal mapping to 2Lh was recovered out of 2,363 chromosomes. Thus, I-R dysgenesis was highly effective in inducing mutations of chromosome 2 heterochromatic genes. The lethal chromosomes were further characterized in complementation assays with EMS-induced point mutations belonging to 13 heterochromatic complementation groups on chromosome 2 (22). The results of this analysis are summarized in Table 2. Twenty-two lethal mutations genetically behaved like point mutations. Interestingly, 8 chromosomes failed to complement more than one of the EMS-induced mutations and thus behaved as heterochromatic rearrangements. For example, IR-1 failed to complement the four most proximal 41Ab, 41 Aa, rl, and 41Ad genes, while IR-11 simultaneously lacked 40Fg and 40Ff loci, yet retained 40Fe, which lies in between 40Fg and 40Ff (23), and thus likely corresponds to an inversion. In addition, IR-45 behaved like a complex rearrangement, rather than a polar deletion. Finally, IR-3, IR-4, IR-23, IR-24, and IR-30, although complementing EMS alleles of all six known 2Rh loci, failed to complement Df(2Rh)M-S28, which removes the distal 41Ae and 41Af loci. The lethal mutations on these chromosomes were rescued by the e97 duplication carrying the entire 2Rh (29). Thus, these chromosomes may carry mutations in genes mapping to the distal portion of 2Rh other than 41Ae and 41Af. We also performed an F1 screen for the recovery of I-R dysgenesis-induced mutations in the lt and rl genes. Charolles (R) females were crossed to Canton-S (I) males, and their dysgenic daughters were subsequently mated to b lt rl cn homozygous males to select lt and rl phenotypes. Two lethal light (ltIR1 and ltIR2) and two vital rl alleles (rlIR1 and rlIR2) were recovered from 15,777 individuals, which corresponds to a frequency of 10−4 for both genes. No heterochromatic mutation was recovered from 19,838 individuals scored in the reciprocal nondysgenic cross.
Table 2.
Genetic analysis of heterochromatic mutations induced by the I-R dysgenesis
| Cross | Lethals | Allelism test |
|---|---|---|
| 1 | l(2Rh)IR1 | 41Ab; 41Aa; rl; 41Ad |
| l(2Rh)IR2 | 41Ad | |
| l(2Rh)IR3 | Lethal with M-S210and | |
| l(2Rh)IR4 | M-S28; vital with e97 | |
| l(2Rh)IR5 | 41Ah | |
| l(2Rh)IR6 | 41Ah | |
| l(2Rh)IR7 | 41Aa; rl | |
| l(2Rh)IR8 | 41Ad | |
| l(2Rh)IR9 | 41Ad | |
| l(2Rh)IR17 | 41Ah | |
| l(2Rh)IR18 | 41Aa | |
| l(2Rh)IR37 | rl | |
| l(2Rh)IR38 | 41Ad | |
| l(2Rh)IR39 | 41Ad | |
| l(2Lh)IR10 | Lethal with Sdlt5and C′ | |
| l(2Lh)IR11 | 40Ff; 40Fg | |
| l(2Lh)IR12 | Semilethal with Sdlt5and C′ | |
| l(2Lh)IR29 | 40Ff; 40Fe; 40Fg | |
| l(2Lh)IR31 | 40Fe | |
| l(2Lh)IR32 | 40Ff | |
| l(2Lh)IR33 | 40Fe; 40Fg | |
| l(2Lh)IR34 | 40Fe | |
| l(2Lh)IR35 | 40Fe; 40Fg | |
| l(2Lh)IR36 | 40Ff; 40Fe; 40Fg; 40Fb | |
| 2 | l(2Rh)IR13 | 41Ae |
| l(2Rh)IR14 | 41Ae | |
| 3 | l(2Rh)IR21 | 41Ad |
| l(2Rh)IR23 | Lethal with M-S210and | |
| l(2Rh)IR24 | M-S28; vital with e97 | |
| l(2Rh)IR27 | rl | |
| l(2Rh)IR30 | Lethal with M-S210and M-S28 | |
| 4 | l(2Rh)IR25 | 41Ae |
| l(2Rh)IR26 | 41Ae | |
| l(2Lh)IR20 | 40Fg | |
| l(2Lh)IR28 | 40Fe; 40Fg | |
| 5 | l(2Rh)IR40 | 41Ad |
| l(2Rh)IR41 | rl | |
| l(2Rh)IR42 | 41Ab; 41Aa; rl; 41Ad | |
| l(2Rh)IR43 | 41Ah | |
| l(2Lh)IR45 | Retains only 40Fc | |
| l(2Rh)IR46 | 41Ad | |
| l(2Rh)IR47 | 41Ad |
Crosses 1 = Ch × C-S; 2 = cn × Cy/Pm; 3 = Ch × M-5; 4 = cn × C-S; and 5 = Ch × b lt rl cn, in which Ch = Charolles and C-S = Canton-S. The following EMS-induced alleles representing 13 genetic loci mapping to the heterochromatin of chromosome 2 were used in complementation tests: 56-8, 56-32, 56-3, 40-7, 56-4, 56-24, and 40-5 for 2Lh and 45-10, 31, 64, 45-17, 34-25, and EMS 45-75 for 2Rh.
Euchromatic and Heterochromatic Lethals Induced by I-R Dysgenesis.
We next performed dysgenic crosses designed to compare the frequency of euchromatic vs. heterochromatic lethals simultaneously induced along chromosome 2. A total of 45 homozygous lethal chromosomes were recovered from 830 chromosomes in I-R dysgenic crosses, while 6 lethal chromosomes were found over 539 chromosomes in nondysgenic controls; the difference in the lethal frequency between experimental and control groups is statistically significant (χ2 = 15.73; P ≤ 0.001). Five lethal chromosomes from the dysgenic crosses (IR-29, IR-36, IR-37, IR-38, and IR-39) were found to carry heterochromatic lethals (Table 2). Among them, IR-29 behaved like a deletion, while IR-36 was likely to correspond to a multibreak rearrangement; the remaining 40 lethals complemented heterochromatic deficiencies of both 2R and 2L, and thus mapped to the euchromatin. None of the 6 lethals recovered from control crosses were heterochromatic; they may have been induced by a basal level of dysgenesis, and/or may correspond to preexisting lethals in Canton-S chromosome 2 that was not isogenized. To summarize, these experiments yielded euchromatic recessive lethals with a frequency close to 4.8 × 10−2, and heterochromatic lethals with a frequency of 6 × 10−3.
Cytological Analysis of I-R Dysgenesis-Induced Heterochromatic Lethals.
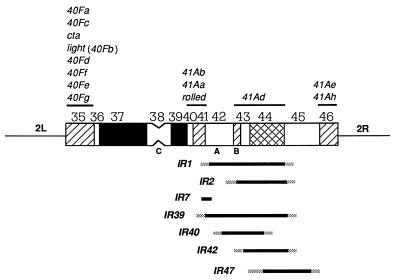
To establish whether structural alterations had occurred in the heterochromatin of I-R-induced lethals, mitotic chromosomes of 39 lines were examined after Hoechst 33258 banding. The cytological data are summarized in the map in Fig. 1. The IR-1, IR-7, and IR-42 lethals carry deletions affecting the 2R heterochromatin, in agreement with the genetic data (Table 2). The deletions occurring in both IR-1 and IR-42 spanned the region included between the proximal part of h41 and h44, while IR-7 lacked a large part of h41. The IR-2, IR-39, IR-40, and IR-47 mutations, which genetically belong to the 41Ad complementation group only, were also found to be deletions: for example, both IR-2 and IR-40 span the region from h42 to h44. In contrast, no cytological abnormality was detected in IR-11, IR-28, IR-29, IR-33, IR-35, IR-36, and IR-45, which genetically behaved as rearrangements (see Table 2), or among the remaining lethals. In conclusion, genetic and cytological analyses show that almost 35% of all analyzed heterochromatic lethals were associated with chromosome rearrangements which mostly corresponded to large heterochromatic deletions. Interestingly, in an independent study about 30% of the I-R-induced euchromatic lethals of the X chromosome were also found to be associated with chromosomal rearrangements (31).
Figure 1.
Cytogenetic map of chromosome 2 heterochromatin, showing the extent of the I-R-induced deficiencies along the heterochromatin of the right arm. Thirteen cytologically distinct regions, designated h35 to h46, can be resolved in chromosome 2 heterochromatin (24). Filled areas represent the Hoechst 33258-bright regions; the shaded boxes represent regions of intermediate fluorescence, and the open boxes are regions of dull fluorescence. Only the heterochromatic regions are shown. Above the diagram are bars indicating the mapping of 13 vital heterochromatic genes. The genes in the left arm of chromosome 2 (2Lh) are indicated from top to bottom according to their left to right mapping along the chromosome. The extent of the I-R-induced deficiencies was determined cytogenetically as described in detail elsewhere (24, 30) and is represented by thick lines (below). 2L, left arm; 2R, right arm; C, centromeric region.
Heterochromatic Recombination Induced by I-R Dysgenesis.
Although heterochromatin does not undergo meiotic recombination with an appreciable frequency, Hirazumi reported recombination between lt and Responder (Rsp) loci following I-R dysgenesis (32). We wished to investigate the possibility that heterochromatic recombination was involved in the generation of lethals. To that aim, we measured the frequency of both heterochromatic recombinants and heterochromatic lethals recovered from the same dysgenic cross involving the wild-type Charolles (R) strain and the b lt rl cn (I) strain (see Materials and Methods for details). The results of these experiments are shown in Tables 1, 3, and 4. The frequency of recombinants between the b and lt markers of 2L and between rl and cn in 2R does not increase in the dysgenic cross compared with nondysgenic control (Table 3). In contrast, the frequency of recombinants between lt and rl approaches 10−2 in the dysgenic experiment and is significantly higher, by nearly two orders of magnitude, than that observed in the control. Among a subset of 50 randomly selected recombinants between lt and rl, one chromosome, designated R1(+ + rl cn) had acquired a lethal allele of the heterochromatic l(2Rh)41Ad gene, which is located distally to rl. Therefore, the frequency of heterochromatic lethals within the group of recombinant chromosomes approached 2 × 10−2 (1/50) and was not significantly different from the 0.6 × 10−2 frequency that we found within the group of chromosomes that had not been selected for recombination (see crosses 7a and 7b in Table 1). When we determined the fraction of heterochromatic recombinants in the group of heterochromatic lethals, we found that two (IR-40 and IR-47) of seven lethals (28%) were recombinants between the rl and cn markers (Table 4). In this case, the frequency of recombinants is significantly higher (χ2 = 11.54; P ≤ 0.001) than that measured in a group of chromosomes that had not been selected for lethality (see Table 3). Both IR-40 and IR-47 were alleles of the 41Ad gene. In addition, among a group of twenty b lt rl + recombinants, one (R31) was found to be a lethal allele of 41Ad. It is intriguing that IR-40, IR-47, and R31 had all independently accumulated a lethal allele of the same l(2Rh)41Ad gene, which maps to the h44 region of 2Rh (see Fig. 1) between rl and cn. Both the IR-40 and IR-47 lethals cytologically corresponded to deletions of 2Rh affecting regions h43–44–45 (Fig. 1), while R31 showed no chromosomal alteration.
Table 3.
I-R dysgenesis-induced recombination between lt and rl
| Cross | No. of parentals b lt rl cn + + + + | No. of recombinants
|
Frequency of recombinants
|
||||
|---|---|---|---|---|---|---|---|
| b + + + + lt rl cn | b lt rl + + + + cn | b lt + + + + rl cn | b and lt | rl and cn | lt and rl | ||
| Exp. | 6,298 | 338 | 126 | 75 | 4.9 × 10−2 | 1.8 × 10−2 | 1 × 10−2 |
| Control | 8,037 | 457 | 138 | 2 | 5.3 × 10−2 | 1.6 × 10−2 | 2.3 × 10−4 |
Exp. = dysgenic crosses; Control = nondysgenic controls.
Table 4.
Recombinants between rl and cn within a group of chromosome 2 heterochromatic lethals
| Lethals | Genotype | Allelism test |
|---|---|---|
| l(2Rh)IR40 | b lt rl + | 41Ad (Df) |
| l(2Rh)IRh41 | b lt rl cn | rl |
| l(2Rh)IRh42 | b lt rl cn | 41Aa; rl; 41Ad (Df) |
| l(2Rh)IRh43 | b lt rl cn | 41Ah |
| l(2Rh)IRh46 | b lt rl cn | 41Ad |
| l(2Rh)IRh47 | + + + cn | 41Ad (Df) |
| l(2Lh)IRh45 | + lt + + | Retains only 40Fc |
IRh41, IRh42, IRh43, and IRh42 are associated with the b lt rl cn chromosomes; IRh40 and IRh47 are associated with the b lt rl + and + + + cn recombinant chromosomes, respectively; IRh45 is a deletion which is likely to be induced on the Charolles wild-type chromosome. (Df) = cytologically detectable deletions.
I Factor Insertion Within the rl Gene.
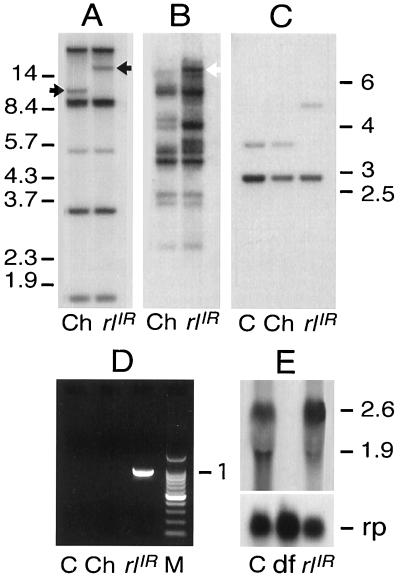
To investigate the molecular nature of I-R dysgenesis-induced heterochromatic mutations, we initially carried out Southern blot hybridization experiments to analyze the rl region in chromosomes carrying the homozygous viable rlIR1 allele in comparison with wild-type parental chromosomes. The rlIR1 allele arose from dysgenic crosses between the Charolles (R) and Canton-S (I) strains. Consistent with the large size of the rl locus (W. H. Biggs III and K. Zavitz, personal communication), several genomic fragments could be identified after digestion with EcoRI, which does not cut within the I factor. The only visible difference in the digestion pattern of the parental rl+ Charolles compared with the mutant strain consisted in the altered mobility of one hybridizing band, which shifted from 10 kb to about 15 kb (Fig. 2A); this difference is consistent with what would be expected from the insertion of a full-length I factor. Reprobing the filter with a 1.5-kb probe containing the I factor 5′ end (Fig. 2B) showed that the newly generated 15-kb band of rlIR1 indeed comigrated with I factor-homologous DNA sequences. Similar results were obtained after digestion with PstI and XhoI (data not shown). A more detailed mapping of the putative I factor insertion was obtained using a probe corresponding to a 500-bp HindIII–DraI fragment, containing exons 1 and 2 of the rl gene (W. H. Biggs III and K. Zavitz, personal communication). Only two bands, of about 3 kb and 3.5 kb respectively, hybridized with this probe in both Canton-S and Charolles rl+ parental strains after EcoRV digestion (Fig. 2C). In the rlIR1 mutant, a novel 5-kb band could be seen, which replaced the parental 3.5-kb fragment. The observed difference in the size of these two bands corresponds to about 1.5 kb, in agreement with the presence of an EcoRV site at position 1570 in the I factor DNA. The rl exons 1 and 2 were found to be 350 and 140 bp long, respectively (W. H. Biggs III and K. Zavitz, personal communication), suggesting that the weaker signal intensity over the 3.5-kb parental band, which shifts to 5 kb in rlIR1, might reflect the hybridization with exon 2 sequences. Together, these results suggest that rlIR1 corresponds to an I factor insertion closely linked to exon 2.
Figure 2.
(A) Southern hybridization of genomic DNA from the Charolles parental strain (Ch) carrying the rl+ allele, and from rlIR1 homozygotes (rlIR), digested with EcoRI and probed with the 1.4-kb rl cDNA probe (30); black arrows indicate the bands whose mobility varies in the rlIR1 mutant and in the Charolles strain. (B) Filters were stripped and reprobed with I sequences; the open arrow indicates I-homologous sequences in the dysgenic mutants that comigrate with rl homologous sequences. (C) Southern hybridization of genomic DNA from Canton-S (C) and Charolles (Ch) parental stains carrying the rl+ allele, and from rlIR1 homozygotes (rlIR), digested with EcoRV and probed with the HindIII–DraI fragment. (D) PCR amplification reaction from genomic DNA of Canton-S (C), Charolles (Ch), and rlIR1 strains. A 982-bp fragment was specifically amplified from genomic DNA of rlIR1 homozygotes. M, molecular weight marker. (E) Northern blot assays of second-instar larvae total RNA from rl+, rlIR1, and Df(2Rh)Rspi1 homozygotes lacking the rl gene. The 1.4-kb rl cDNA probe reveals two major transcripts of 2.6 kb and 1.9 kb, both absent in larvae homozygous for Df(2Rh)Rspi1. The same filter was reprobed with the rp49 ribosomal protein gene probe as a loading control.
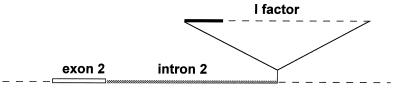
To learn more about the nature of the rlIR1 mutation, we used different pairs of oligonucleotide primers in PCR amplification reactions from genomic DNA of Charolles, Canton-S, and rlIR1 strains. Specific sets of primers were synthesized from different regions of rl exon 2 and from the I factor (see Materials and Methods for details), to amplify possible integration products of I within rl DNA, as suggested by the Southern analysis. Using the rl3 and I5R combination of primers, directed respectively downstream of rl exon 2 and forward from the I factor 5′ end, we could specifically amplify a 982-bp fragment from genomic DNA of rlIR1 homozygotes (Fig. 2D). Sequence analysis showed that the PCR fragment indeed contained 125 bp of rl exon 2, followed by a 394-bp region of unidentified homology, which likely corresponds to sequences of rl intron 2, and by 462 bp, representing the terminal part of the I factor 5′ end (GenBank accession no. Y11299). These results clearly show that the rlIR1 allele is associated with a de novo insertion of the I factor into a rl intron, 400 bp downstream of exon 2 (Fig. 3).
Figure 3.
Schematic representation of the I factor insertion in the rl gene. The white and shaded lines represent 125 bp of the rl exon 2, and 394 bp of the rl intron 2 respectively; the filled line represents the 5′ end of the I factor. Dashed lines represent rlIR1 genomic sequences and I factor sequences that were not included in the 982-bp PCR fragment. The I factor insert is not in scale.
Consistent with the findings that the I factor had been inserted in a intronic region of the rl gene, no detectable difference was observed in the size of mRNA transcripts homologous to the rl cDNA probe from the parental and rlIR1 strains (Fig. 2E), although it remains possible that the I insertion caused abnormalities in the splicing and/or processing of the RNA transcripts.
DISCUSSION
In this work, we have investigated whether chromosome 2 heterochromatic genes of D. melanogaster are mutable after I-R hybrid dysgenesis, where transposition of the LINE-like I factors is activated. An interesting outcome of our studies is the high overall frequency of lethal mutations mapping to chromosome 2 heterochromatin in dysgenic crosses, approaching 10−2 (Table 1). In particular, the 41Ad gene is the most highly mutable heterochromatic gene of chromosome 2, and it exhibits an overall frequency of mutation (1.5 × 10−3) which is one order of magnitude greater than that calculated for the most mutable euchromatic genes in I-R dysgenic crosses (17, 21). In addition, along chromosome 2, which carries at least 2,000 genes, euchromatic mutations were recovered with a frequency close to 5 × 10−2; in the same experiments, heterochromatic mutations were recovered with a frequency of about 0.6 × 10−2: thus, the euchromatin is only 8-fold more susceptible to mutate to recessive lethal than heterochromatin, despite its 150-fold enrichment in genetic functions. It follows that heterochromatic genes are about 20-fold more mutable than euchromatic genes in chromosome 2. About 35% of the analyzed heterochromatic lethals were associated with rearrangements which mostly correspond to large deletions spanning several megabases of DNA. The remaining lethals affecting individual loci may correspond to I factor insertions into the heterochromatic genes, as shown in the case of the rlIR1 mutation (Figs. 2 and 3); in other cases, the occurrence of small lesions beyond the cytological level of resolution cannot be ruled out.
We have also found that I-R dysgenesis induces 1% recombination between the lt and rl genes in chromosome 2 heterochromatin, a value which is nearly two orders of magnitude higher than observed in nondysgenic controls. However, recombination between lt and rl does not appear to be responsible for the generation of lethals. In contrast, recombination between rl and cn is indeed involved in the specific generation of lethal alleles of the 41Ad gene (Table 4). Interestingly, two of them (IR40 and IR47) correspond to cytologically detectable deletions of 2Rh (Fig. 1) which might result from unequal exchanges. I-R dysgenesis was previously reported to induce both insertional mutations and chromosomal rearrangements in the X chromosome euchromatin (21, 31, 33). In most examined cases, I factor sequences were found to map at the breakpoints of the I-R-induced rearrangements, which were suggested to result either from recombination between transposing copies of I at the time of integration or, alternatively, from a target exchange between I factors. According to the latter model, the same element would join two nicks generated in nonhomologous chromosomal regions.
Non-LTR retrotransposons have been shown to integrate at specific heterochromatic sites in the Drosophila genome; for example, the R1 and R2 elements insert into specific sequences of the 28S rRNA genes (15). The Het-A and TART elements also specifically transpose with high frequency to broken chromosomal ends, which are regarded as being heterochromatic (18). The high frequency of mutations reported in the present study suggests that single-copy heterochromatic genes of chromosome 2 are a preferential target for I factor retrotransposition, which in turn would produce either insertional events or chromosomal rearrangements. This conclusion conflicts with the view that constitutive heterochromatin is mostly insensitive to the mutational effects caused by TEs (10).
Because TEs induce chromosome rearrangements in both natural and laboratory populations of Drosophila, they have been proposed to shape the genome structure (34). In addition, TEs may drive the concerted evolution of repetitive DNA sequences (35). It is tempting to speculate that the heterochromatic variability generated by both mutations and recombination events during I-R dysgenesis might have played a role in the evolution of Drosophila heterochromatin. Polymorphisms of constitutive heterochromatin are known to occur in natural populations of higher eukaryotes, including Drosophila and humans (36–38). Contraction or expansion of heterochromatic regions, such as those depicted in the Rsp locus (30, 39), are thought to result from unequal exchanges between satellite DNA repeats. It is not unreasonable to suggest that these events could also be stimulated by I-R dysgenesis-induced heterochromatic recombination. Interestingly, L1 elements were suggested to be involved in the generation of heterochromatic variability in human chromosomes (40). Further investigations on the interactions between LINEs and heterochromatic regions will be necessary to further clarify the impact of TEs upon heterochromatin evolution in eukaryotes. Finally, insertional mutagenesis with marked I factor might represent a powerful tool for molecular dissection of heterochromatin and cloning of heterochromatic genes.
Acknowledgments
We are grateful to N. Junakovic, M. Gatti, P. Lavia, A. Pelisson, and F. Spirito for helpful discussions. We also thank A. Pelisson and C. Vaury for generously providing reactive and inducer strains; C. Di Franco, E. Hafen, and L. Zypursky for the gift of DNA clones; I. Busseau, N. Hollers, and E. Hafen for the gift of DNA primers; and W.H. Biggs III and K. Zavitz for communicating results before publication.
ABBREVIATIONS
- TE
transposable element
- LTR
long terminal repeat
- LINE
long interspersed nuclear element
- EMS
ethyl methanesulfonate
References
- 1.Finnegan D J. Trends Genet. 1989;5:103–107. doi: 10.1016/0168-9525(89)90039-5. [DOI] [PubMed] [Google Scholar]
- 2.Grimaldi G, Singer M F. Nucleic Acids Res. 1983;11:321–338. doi: 10.1093/nar/11.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young B S, Passion A, Traverse K L, French C, Pardue M L. Cell. 1983;34:85–94. doi: 10.1016/0092-8674(83)90138-1. [DOI] [PubMed] [Google Scholar]
- 4.Kuff E L, Fewell J E, Lueders K K, Di Paolo J A, Amsbaugh S C, Popescu N C. Chromosoma. 1986;93:213–219. doi: 10.1007/BF00292740. [DOI] [PubMed] [Google Scholar]
- 5.Taruscio D, Manuelidis L. Chromosoma. 1991;101:141–156. doi: 10.1007/BF00355364. [DOI] [PubMed] [Google Scholar]
- 6.Steinmann M, Steinmann S. Proc Natl Acad Sci USA. 1993;89:7591–7595. doi: 10.1073/pnas.89.16.7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaury C, Bucheton A, Pelisson A. Chromosoma. 1989;98:215–224. doi: 10.1007/BF00329686. [DOI] [PubMed] [Google Scholar]
- 8.Pimpinelli S, Berloco M, Fanti L, Dimitri P, Bonaccorsi S, Marchetti E, Caizzi R, Caggese C, Gatti M. Proc Natl Acad Sci USA. 1995;92:3804–3808. doi: 10.1073/pnas.92.9.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lansman R A, Shade R O, Grigliatti T A, Brock H W. Proc Natl Acad Sci USA. 1987;84:6491–6495. doi: 10.1073/pnas.84.18.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charlesworth B, Jarne P, Assimacopoulos S. Genet Res. 1994;64:183–197. doi: 10.1017/s0016672300032845. [DOI] [PubMed] [Google Scholar]
- 11.Gatti M, Pimpinelli S. Annu Rev Genet. 1992;26:239–275. doi: 10.1146/annurev.ge.26.120192.001323. [DOI] [PubMed] [Google Scholar]
- 12.Devlin R H, Bingham B, Wakimoto B T. Genetics. 1990;125:129–140. doi: 10.1093/genetics/125.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang P, Spradling A. Proc Natl Acad Sci USA. 1994;91:3539–3543. doi: 10.1073/pnas.91.9.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roseman R R, Johnson E A, Rodesch C K, Bjerke M, Nagoshi R N, Geyer P K. Genetics. 1995;141:1061–1074. doi: 10.1093/genetics/141.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heickbush T H. New Biol. 1992;4:430–440. [PubMed] [Google Scholar]
- 16.Hutchinson C A, Hardies S C, Loeb D L, Berg D E, Howe M M, Shehee W R, Edgell M H. In: Mobile DNA. Berg D E, Howe M M, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 593–617. [Google Scholar]
- 17.Finnegan D J. In: Mobile DNA. Berg D E, Howe M M, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 503–517. [Google Scholar]
- 18.Pardue M L, Danilevskaya O N, Lowenhaupt K, Slot F, Traverse K L. Trends Genet. 1996;12:48–52. doi: 10.1016/0168-9525(96)81399-0. [DOI] [PubMed] [Google Scholar]
- 19.Picard G, L’Heritier P. Drosophila Inf Serv. 1971;46:54. [Google Scholar]
- 20.Bucheton A, Paro R, Sang H M, Pelisson A, Finnegan D J. Cell. 1984;38:153–163. doi: 10.1016/0092-8674(84)90536-1. [DOI] [PubMed] [Google Scholar]
- 21.Busseau I, Chaboissier M C, Pelisson A, Bucheton A. Genetica. 1994;93:101–116. doi: 10.1007/BF01435243. [DOI] [PubMed] [Google Scholar]
- 22.Hilliker A J. Genetics. 1976;83:765–782. doi: 10.1093/genetics/83.4.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wakimoto B T, Hearn M G. Genetics. 1990;125:141–154. doi: 10.1093/genetics/125.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dimitri P. Genetics. 1991;127:553–564. doi: 10.1093/genetics/127.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiler K S, Wakimoto B T. Annu Rev Genet. 1995;29:577–605. doi: 10.1146/annurev.ge.29.120195.003045. [DOI] [PubMed] [Google Scholar]
- 26.Lindsley D L, Zimm G L. The Genome of Drosophila melanogaster. New York: Academic; 1992. [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 28.Biggs, H. W., Zavitz, H. K., Dikinson, B., Van Der Straten, A. & Brunner, D. EMBO J. 13, 1628–1635. [DOI] [PMC free article] [PubMed]
- 29.Brittnacher J G, Ganetzky B. Genetics. 1989;121:739–750. doi: 10.1093/genetics/121.4.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pimpinelli S, Dimitri P. Genetics. 1989;121:765–772. doi: 10.1093/genetics/121.4.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Proust J, Prudhommeau C, Ladeveze V, Gotteland M, Fontyne-Blanchard M C. Mutat Res. 1992;268:265–285. doi: 10.1016/0027-5107(92)90233-r. [DOI] [PubMed] [Google Scholar]
- 32.Hiraizumi Y. Genetics. 1981;98:105–114. doi: 10.1093/genetics/98.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Busseau I, Pelisson A, Bucheton A. Mol Gen Genet. 1989;218:222–228. doi: 10.1007/BF00331272. [DOI] [PubMed] [Google Scholar]
- 34.Lim J K, Simmons M J. BioEssays. 1994;16:269–275. doi: 10.1002/bies.950160410. [DOI] [PubMed] [Google Scholar]
- 35.Stewart-Thompson D, Karpen G H, Spradling A C. Proc Natl Acad Sci USA. 1994;91:9042–9046. doi: 10.1073/pnas.91.19.9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baimai W. Genetics. 1977;85:85–93. [Google Scholar]
- 37.Halfer C. Chromosoma. 1981;84:195–206. doi: 10.1007/BF00399131. [DOI] [PubMed] [Google Scholar]
- 38.Craig-Holmes A P, Shaw M W. Science. 1971;174:702–704. doi: 10.1126/science.174.4010.702. [DOI] [PubMed] [Google Scholar]
- 39.Wu C-I, Lyttle T W, Wu M-L, Lin G-F. Cell. 1988;54:179–189. doi: 10.1016/0092-8674(88)90550-8. [DOI] [PubMed] [Google Scholar]
- 40.Marcais B, Charlieu J P, Allain B, Brun E, Bellis M, Roizes G. J Mol Evol. 1991;33:42–48. doi: 10.1007/BF02100194. [DOI] [PubMed] [Google Scholar]