Abstract
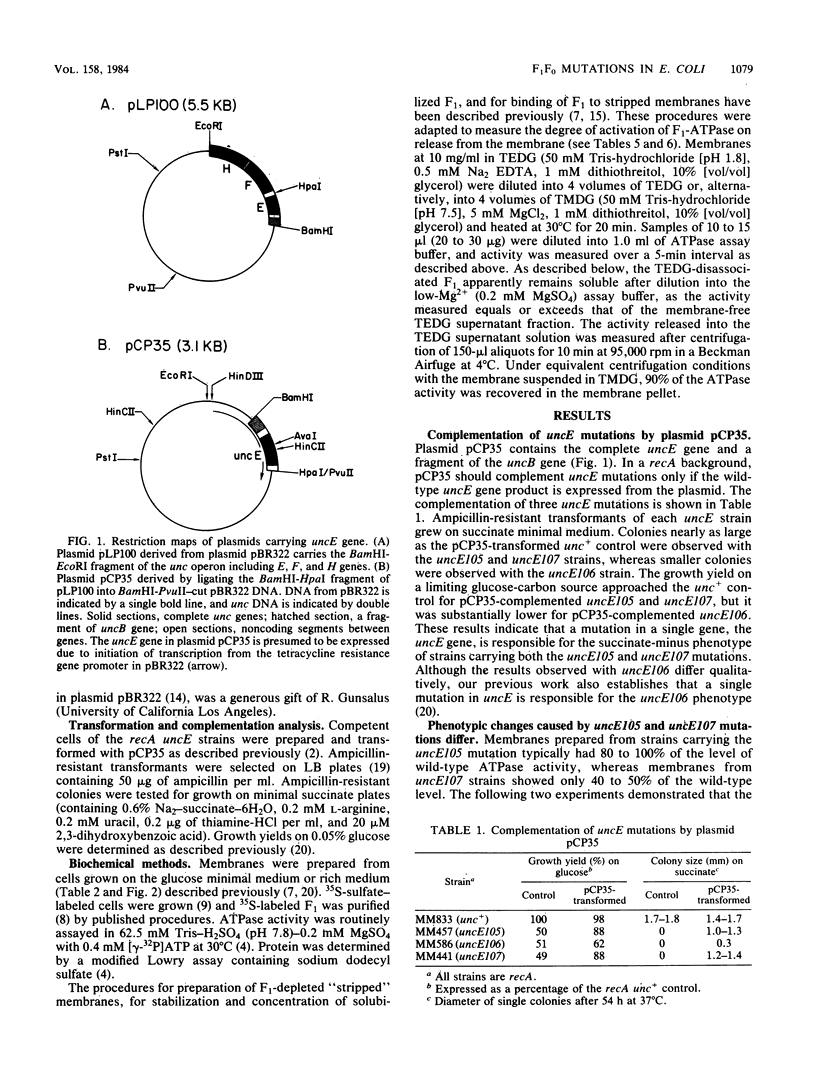
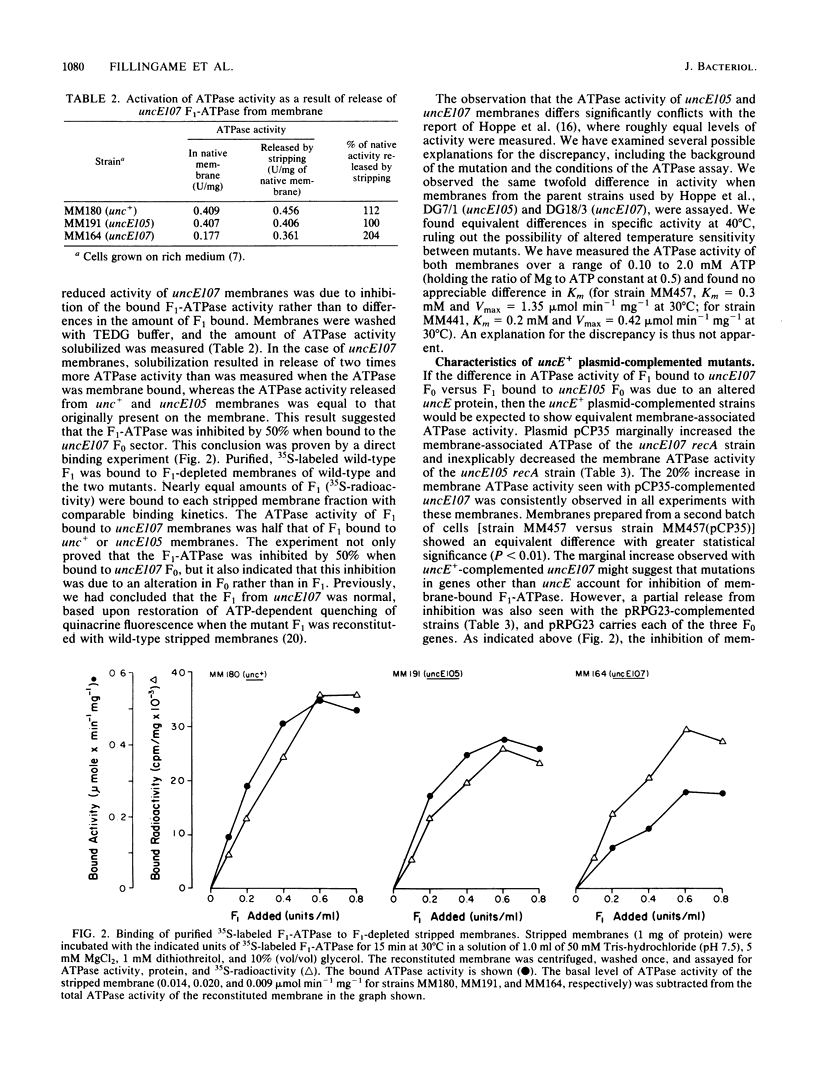
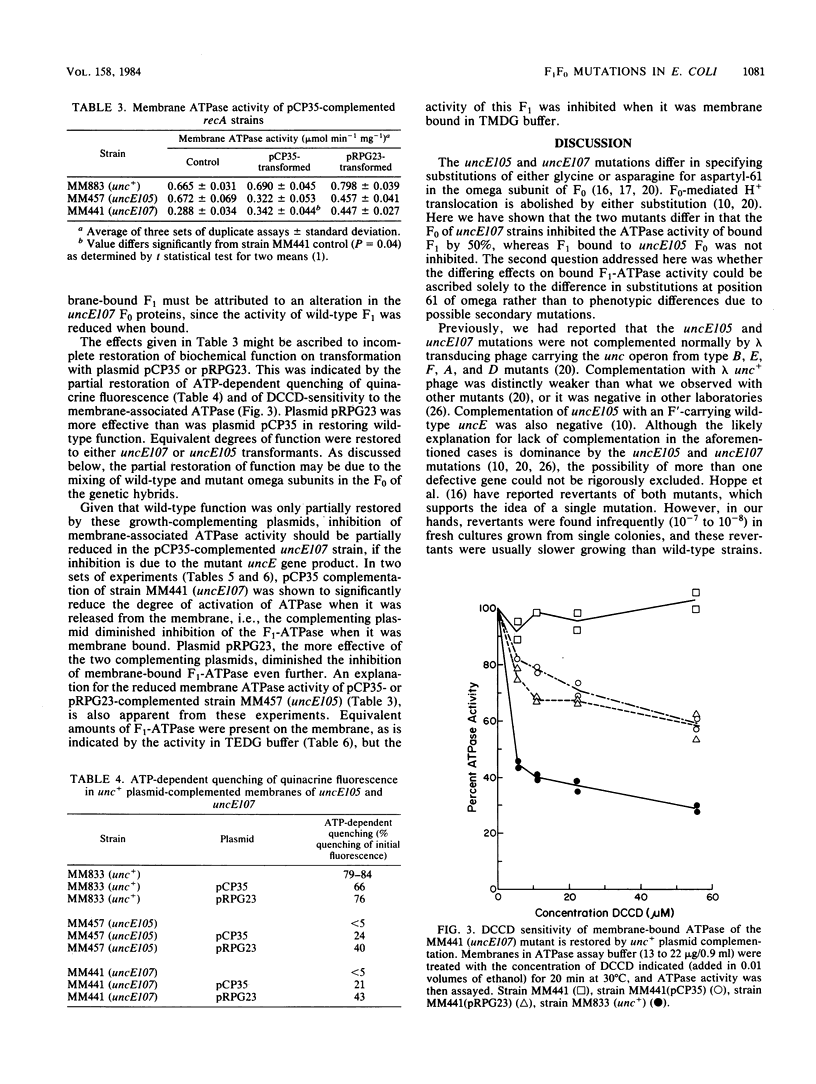
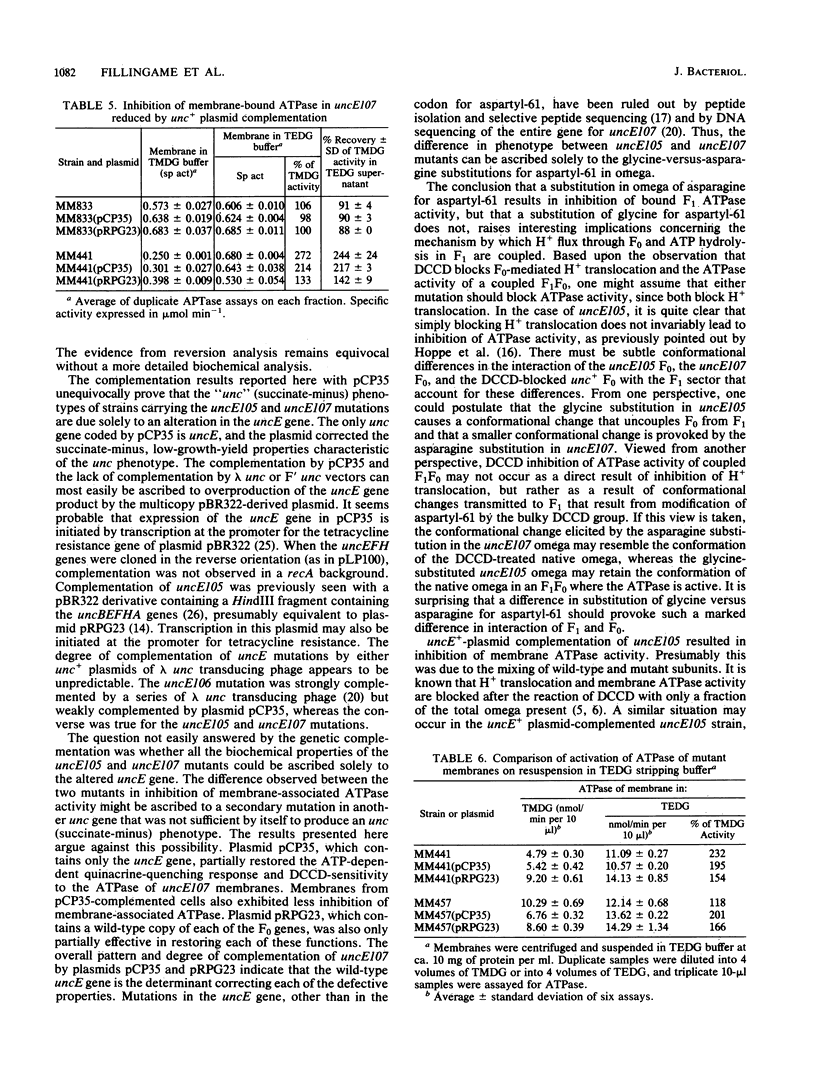
Mutations in the H+-translocating ATPase complex (F1F0) of Escherichia coli have been described in which aspartyl-61 of the omega subunit ( uncE protein) is substituted by either glycine ( uncE105 ) or asparagine ( uncE107 ). Either substitution blocks the H+-translocation activity of the F0 sector of the complex. Here we report a difference in the effects of the two substitutions on the coupled ATPase activity of F1 bound to F0. Wild-type F1 was bound to the F0 of either mutant with affinities comparable to wild-type. The ATPase activity of F1 bound to uncE107 F0 was inhibited by 50%, whereas that bound to uncE105 F0 was not inhibited. Complementation studies with a pBR322-derived plasmid that carried the E gene of the unc operon only indicated that a single mutation in the host strain was responsible for the respective phenotypes. In mutants complemented by the uncE + plasmid, restoration of wild-type biochemical properties was only partial and may be attributed to a mixing of wild-type and mutant omega subunits in a hybrid F0 complex. The activity of membrane-bound F1 was less inhibited in the uncE +/ uncE107 hybrid. Paradoxically, complementation of uncE105 by the uncE + plasmid resulted in substantial inhibition of the activity of membrane-bound F1. The results indicate that a glycine-versus-asparagine substitution for aspartyl-61 must lead to altered conformations of omega and that these differences in conformation are important in the coupling between the F0 and F1 sectors of the complex.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- Downie J. A., Gibson F., Cox G. B. Membrane adenosine triphosphatases of prokaryotic cells. Annu Rev Biochem. 1979;48:103–131. doi: 10.1146/annurev.bi.48.070179.000535. [DOI] [PubMed] [Google Scholar]
- Fillingame R. H. Identification of the dicyclohexylcarbodiimide-reactive protein component of the adenosine 5'-triphosphate energy-transducing system of Escherichia coli. J Bacteriol. 1975 Nov;124(2):870–883. doi: 10.1128/jb.124.2.870-883.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingame R. H., Mosher M. E., Negrin R. S., Peters L. K. H+-ATPase of Escherichia coli uncB402 mutation leads to loss of chi subunit of subunit of F0 sector. J Biol Chem. 1983 Jan 10;258(1):604–609. [PubMed] [Google Scholar]
- Fillingame R. H. Purification of the carbodiimide-reactive protein component of the ATP energy-transducing system of Escherichia coli. J Biol Chem. 1976 Nov 10;251(21):6630–6637. [PubMed] [Google Scholar]
- Foster D. L., Fillingame R. H. Energy-transducing H+-ATPase of Escherichia coli. Purification, reconstitution, and subunit composition. J Biol Chem. 1979 Sep 10;254(17):8230–8236. [PubMed] [Google Scholar]
- Foster D. L., Fillingame R. H. Stoichiometry of subunits in the H+-ATPase complex of Escherichia coli. J Biol Chem. 1982 Feb 25;257(4):2009–2015. [PubMed] [Google Scholar]
- Friedl P., Friedl C., Schairer H. U. F0 of Escherichia coli ATP-synthase containing mutant and wild-type carbodiimide-binging proteins is impaired in H+ -conduction. FEBS Lett. 1980 Oct 6;119(2):254–256. doi: 10.1016/0014-5793(80)80265-1. [DOI] [PubMed] [Google Scholar]
- Futai M., Kanazawa H. Structure and function of proton-translocating adenosine triphosphatase (F0F1): biochemical and molecular biological approaches. Microbiol Rev. 1983 Sep;47(3):285–312. doi: 10.1128/mr.47.3.285-312.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson F. Biochemical and genetic studies on the assembly and function of the F1-F0 adenosine triphosphatase of Escherichia coli. Biochem Soc Trans. 1983 Jun;11(3):229–240. doi: 10.1042/bst0110229. [DOI] [PubMed] [Google Scholar]
- Gibson F., Cox G. B., Downie J. A., Radik J. Partial diploids of Escherichia coli carrying normal and mutant alleles affecting oxidative phosphorylation. Biochem J. 1977 Mar 15;162(3):665–670. doi: 10.1042/bj1620665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus R. P., Brusilow W. S., Simoni R. D. Gene order and gene-polypeptide relationships of the proton-translocating ATPase operon (unc) of Escherichia coli. Proc Natl Acad Sci U S A. 1982 Jan;79(2):320–324. doi: 10.1073/pnas.79.2.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermolin J., Gallant J., Fillingame R. H. Topology, organization, and function of the psi subunit in the F0 sector of the H+-ATPase of Escherichia coli. J Biol Chem. 1983 Dec 10;258(23):14550–14555. [PubMed] [Google Scholar]
- Hoppe J., Schairer H. U., Friedl P., Sebald W. An Asp-Asn substitution in the proteolipid subunit of the ATP-synthase from Escherichia coli leads to a non-functional proton channel. FEBS Lett. 1982 Aug 16;145(1):21–29. doi: 10.1016/0014-5793(82)81198-8. [DOI] [PubMed] [Google Scholar]
- Hoppe J., Schairer H. U., Sebald W. The proteolipid of a mutant ATPase from Escherichia coli defective in H+-conduction contains a glycine instead of the carbodiimide-reactive aspartyl residue. FEBS Lett. 1980 Jan 1;109(1):107–111. doi: 10.1016/0014-5793(80)81321-4. [DOI] [PubMed] [Google Scholar]
- Mosher M. E., Peters L. K., Fillingame R. H. Use of lambda unc transducing bacteriophages in genetic and biochemical characterization of H+-ATPase mutants of Escherichia coli. J Bacteriol. 1983 Dec;156(3):1078–1092. doi: 10.1128/jb.156.3.1078-1092.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrin R. S., Foster D. L., Fillingame R. H. Energy-transducing H+-ATPase of Escherichia coli. Reconstitution of proton translocation activity of the intrinsic membrane sector. J Biol Chem. 1980 Jun 25;255(12):5643–5648. [PubMed] [Google Scholar]
- Schairer H. U., Friedl P., Schmid B. I., Vogel G. The use of several energy-coupling reactions in characterizing mutants of Escherichia coli K12 defective in oxidative phosphorylation. Eur J Biochem. 1976 Jul 1;66(2):257–268. doi: 10.1111/j.1432-1033.1976.tb10515.x. [DOI] [PubMed] [Google Scholar]
- Senior A. E., Wise J. G. The proton-ATPase of bacteria and mitochondria. J Membr Biol. 1983;73(2):105–124. doi: 10.1007/BF01870434. [DOI] [PubMed] [Google Scholar]
- Stüber D., Bujard H. Organization of transcriptional signals in plasmids pBR322 and pACYC184. Proc Natl Acad Sci U S A. 1981 Jan;78(1):167–171. doi: 10.1073/pnas.78.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura F., Kanzawa H., Tsuchiya T., Futai M. Structural gene coding for the dicyclohexylcarbodiimide-binding protein of the protein-translocating ATPase from Escherichia coli. Locus of the gene in the F1--F0 gene cluster. FEBS Lett. 1981 May 5;127(1):48–52. doi: 10.1016/0014-5793(81)80338-9. [DOI] [PubMed] [Google Scholar]


