Abstract
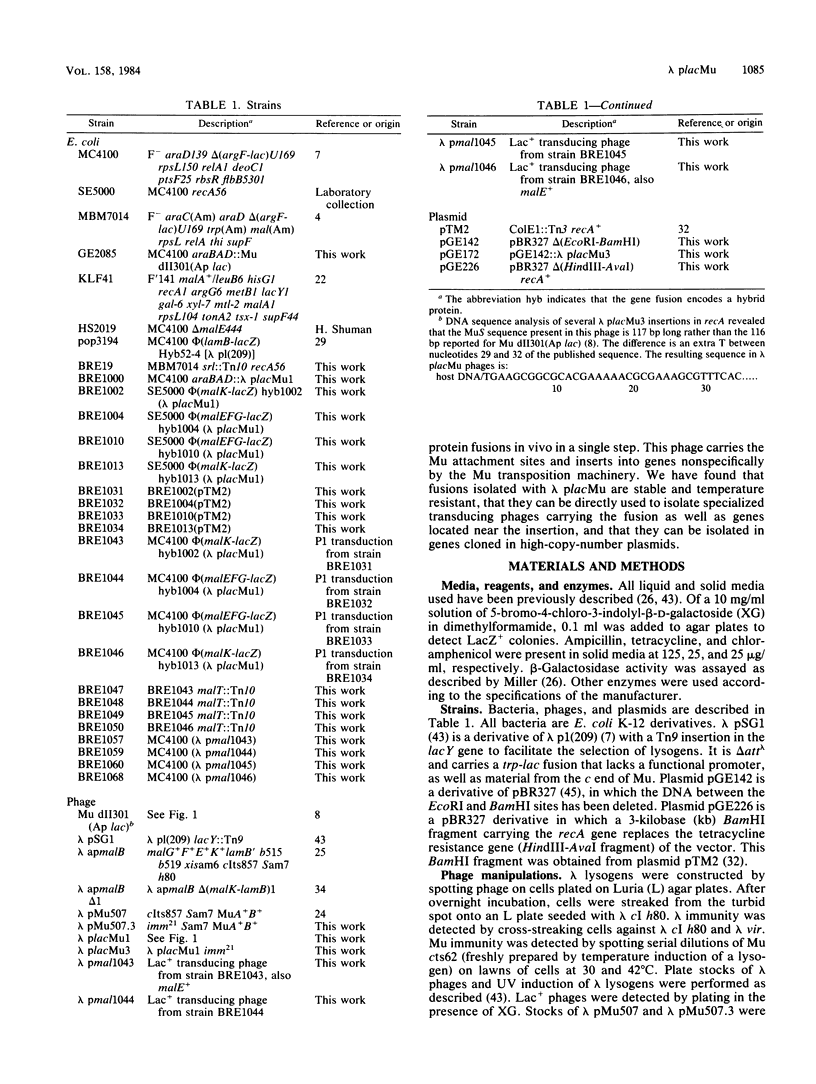
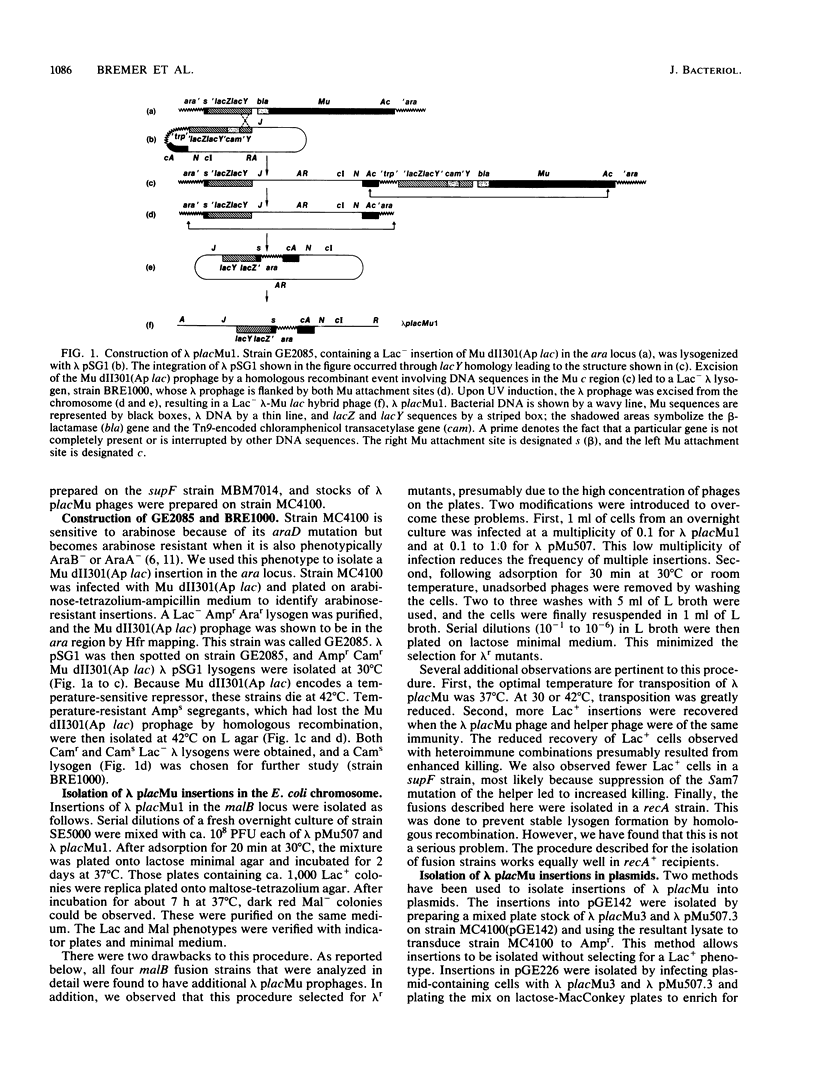
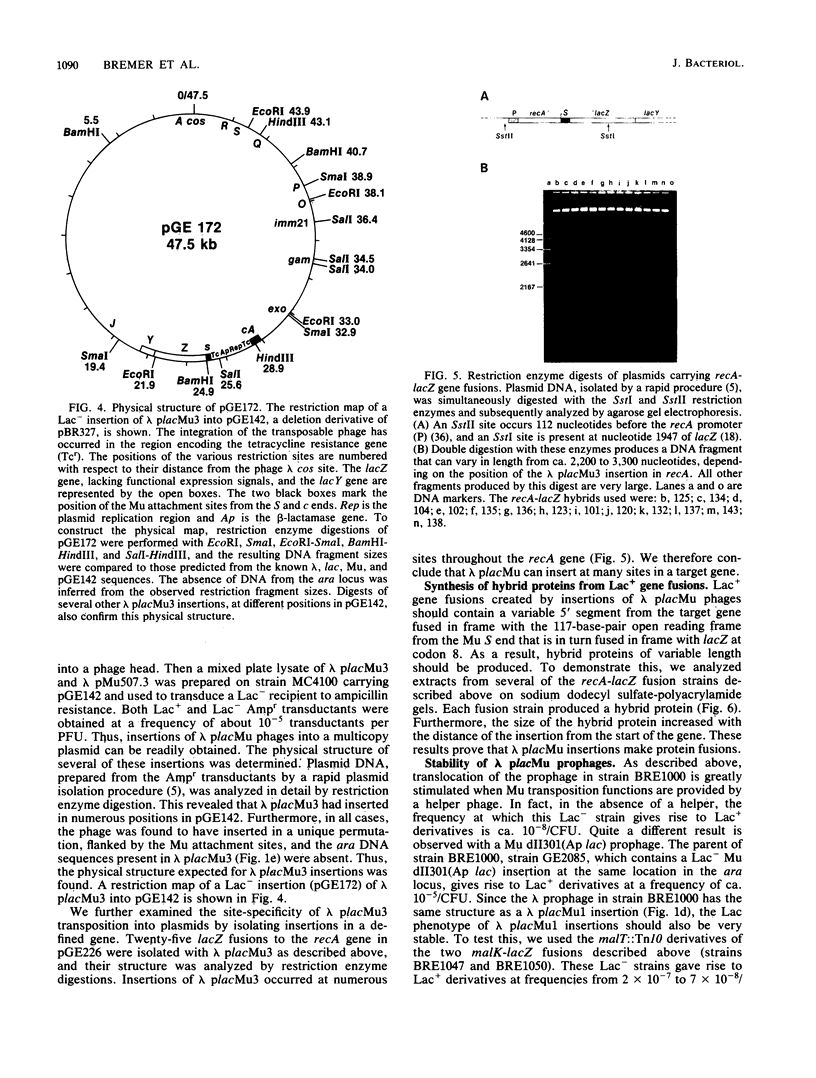
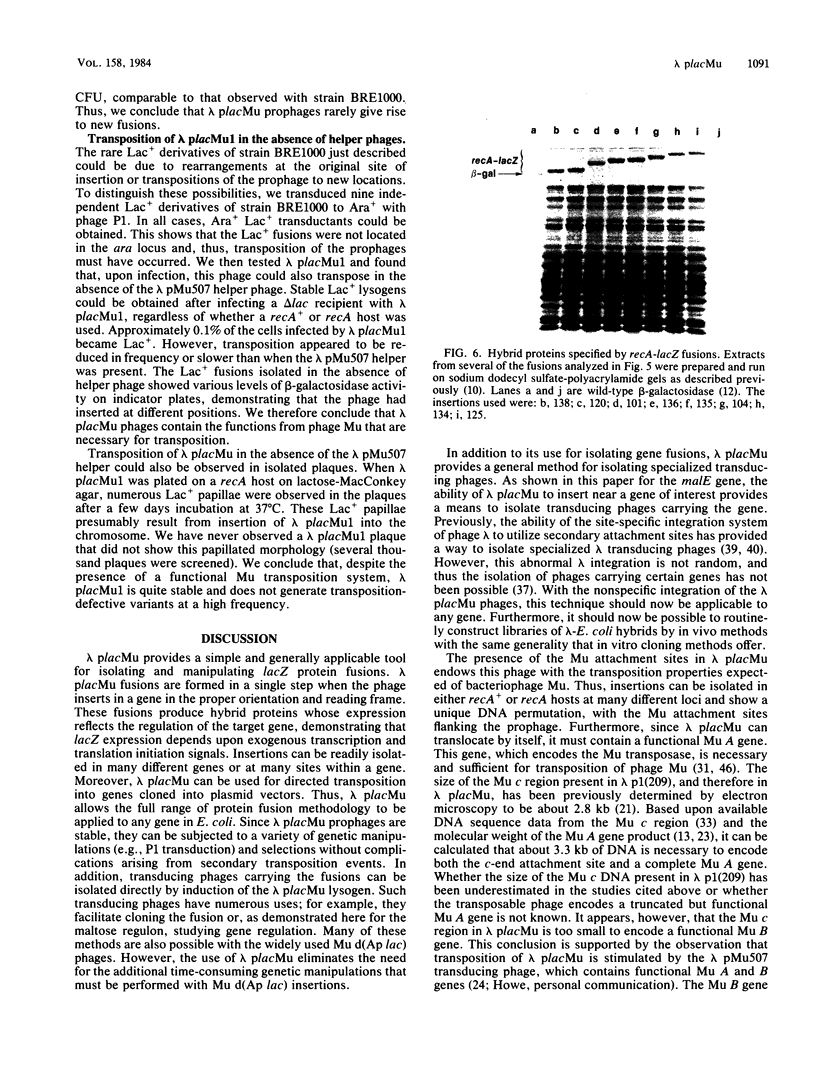
We isolated a plaque-forming derivative of phage lambda, lambda placMu1 , that contains sequences from bacteriophage Mu enabling it to integrate into the Escherichia coli chromosome by means of the Mu transposition system. The Mu DNA carried by this phage includes both attachment sites as well as the cI, ner (cII), and A genes. Lambda placMu1 also contains the lacZ gene, deleted for its transcription and translation initiation signals, and the lacY gene of E. coli, positioned next to the terminal 117 base pairs from the S end of Mu. Because this terminal Mu sequence is an open reading frame fused in frame to lacZ, the phage can create lacZ protein fusions in a single step when it integrates into a target gene in the proper orientation and reading frame. To demonstrate the use of this phage, we isolated lacZ fusions to the malB locus. These showed the phenotypes and regulation expected for malB fusions and could be used to isolate specialized transducing phages carrying the entire gene fusion as well as an adjacent gene (malE). They were found to be genetically stable and rarely (less than 10(-7] gave rise to secondary Lac+ insertions. We also isolated insertions into high-copy-number plasmids. The physical structure of these phage-plasmid hybrids was that expected from a Mu-dependent insertion event, with the lambda placMu prophage flanked by the Mu attachment sites. Lac+ insertions into a cloned recA gene were found at numerous positions and produced hybrid proteins whose sizes were correlated with the position of the fusions in recA.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BECKWITH J. R. A DELETION ANALYSIS OF THE LAC OPERATOR REGION IN ESCHERICHIA COLI. J Mol Biol. 1964 Mar;8:427–430. doi: 10.1016/s0022-2836(64)80206-0. [DOI] [PubMed] [Google Scholar]
- Baker T. A., Howe M. M., Gross C. A. Mu dX, a derivative of Mu d1 (lac Apr) which makes stable lacZ fusions at high temperature. J Bacteriol. 1983 Nov;156(2):970–974. doi: 10.1128/jb.156.2.970-974.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman M. L., Beckwith J. Fusions of the lac operon to the transfer RNA gene tyrT of Escherichia coli. J Mol Biol. 1979 May 25;130(3):285–301. doi: 10.1016/0022-2836(79)90542-4. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Chou J. In vivo formation of gene fusions encoding hybrid beta-galactosidase proteins in one step with a transposable Mu-lac transducing phage. Proc Natl Acad Sci U S A. 1984 Jan;81(2):535–539. doi: 10.1073/pnas.81.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J. Fusion of the Escherichia coli lac genes to the ara promoter: a general technique using bacteriophage Mu-1 insertions. Proc Natl Acad Sci U S A. 1975 Mar;72(3):809–813. doi: 10.1073/pnas.72.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976 Jul 5;104(3):541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- ENGLESBERG E., ANDERSON R. L., WEINBERG R., LEE N., HOFFEE P., HUTTENHAUER G., BOYER H. L-Arabinose-sensitive, L-ribulose 5-phosphate 4-epimerase-deficient mutants of Escherichia coli. J Bacteriol. 1962 Jul;84:137–146. doi: 10.1128/jb.84.1.137-146.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emr S. D., Silhavy T. J. Mutations affecting localization of an Escherichia coli outer membrane protein, the bacteriophage lambda receptor. J Mol Biol. 1980 Jul 25;141(1):63–90. doi: 10.1016/s0022-2836(80)80029-5. [DOI] [PubMed] [Google Scholar]
- Fowler A. V., Zabin I. Amino acid sequence of beta-galactosidase. XI. Peptide ordering procedures and the complete sequence. J Biol Chem. 1978 Aug 10;253(15):5521–5525. [PubMed] [Google Scholar]
- Giphart-Gassler M., Reeve J., van de Putte P. Polypeptides encoded by the early region of bacteriophage Mu synthesized in minicells of Escherichia coli. J Mol Biol. 1981 Jan 5;145(1):165–191. doi: 10.1016/0022-2836(81)90339-9. [DOI] [PubMed] [Google Scholar]
- Gowrishankar J., Pittard J. Construction from Mu d1 (lac Apr) lysogens of lambda bacteriophage bearing promoter-lac fusions: isolation of lambda ppheA-lac. J Bacteriol. 1982 Jun;150(3):1122–1129. doi: 10.1128/jb.150.3.1122-1129.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshey R. M. Switch in the transposition products of Mu DNA mediated by proteins: Cointegrates versus simple insertions. Proc Natl Acad Sci U S A. 1983 Apr;80(7):2012–2016. doi: 10.1073/pnas.80.7.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe M. M., Schumm J. W. Transposition of bacteriophage Mu: properties of lambda phages containing both ends of Mu. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):337–346. doi: 10.1101/sqb.1981.045.01.047. [DOI] [PubMed] [Google Scholar]
- Kalnins A., Otto K., Rüther U., Müller-Hill B. Sequence of the lacZ gene of Escherichia coli. EMBO J. 1983;2(4):593–597. doi: 10.1002/j.1460-2075.1983.tb01468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamp D., Kahmann R. Two pathways in bacteriophage Mu transposition? Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):329–336. doi: 10.1101/sqb.1981.045.01.046. [DOI] [PubMed] [Google Scholar]
- Komeda Y., Iino T. Regulation of expression of the flagellin gene (hag) in Escherichia coli K-12: analysis of hag-lac gene fusions. J Bacteriol. 1979 Sep;139(3):721–729. doi: 10.1128/jb.139.3.721-729.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leathers T. D., Noti J., Umbarger H. E. Physical characterization of ilv-lac fusions. J Bacteriol. 1979 Oct;140(1):251–260. doi: 10.1128/jb.140.1.251-260.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low K. B. Escherichia coli K-12 F-prime factors, old and new. Bacteriol Rev. 1972 Dec;36(4):587–607. doi: 10.1128/br.36.4.587-607.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magazin M., Allet B. Synthesis of bacteriophage Mu proteins in vitro. Virology. 1978 Mar;85(1):84–97. doi: 10.1016/0042-6822(78)90413-0. [DOI] [PubMed] [Google Scholar]
- Magazin M., Howe M., Allet B. Partial correlation of the genetic and physical maps of bacteriophage Mu. Virology. 1977 Apr;77(2):677–688. doi: 10.1016/0042-6822(77)90491-3. [DOI] [PubMed] [Google Scholar]
- Marchal C., Greenblatt J., Hofnung M. malB region in Escherichia coli K-12: specialized transducing bacteriophages and first restriction map. J Bacteriol. 1978 Dec;136(3):1109–1119. doi: 10.1128/jb.136.3.1109-1119.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H., Reznikoff W. S., Silverstone A. E., Ippen K., Signer E. R., Beckwith J. R. Fusions of the lac and trp Regions of the Escherichia coli Chromosome. J Bacteriol. 1970 Dec;104(3):1273–1279. doi: 10.1128/jb.104.3.1273-1279.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D. H., Reznikoff W. S., Beckwith J. R. Genetic fusions defining trp and lac operon regulatory elements. J Mol Biol. 1975 Apr 15;93(3):331–350. doi: 10.1016/0022-2836(75)90281-8. [DOI] [PubMed] [Google Scholar]
- Moreno F., Fowler A. V., Hall M., Silhavy T. J., Zabin I., Schwartz M. A signal sequence is not sufficient to lead beta-galactosidase out of the cytoplasm. Nature. 1980 Jul 24;286(5771):356–359. doi: 10.1038/286356a0. [DOI] [PubMed] [Google Scholar]
- Müller-Hill B., Kania J. Lac repressor can be fused to beta-galactosidase. Nature. 1974 Jun 7;249(457):561–563. doi: 10.1038/249561a0. [DOI] [PubMed] [Google Scholar]
- Priess H., Kamp D., Kahmann R., Bräuer B., Delius H. Nucleotide sequence of the immunity region of bacteriophage Mu. Mol Gen Genet. 1982;186(3):315–321. doi: 10.1007/BF00729448. [DOI] [PubMed] [Google Scholar]
- Raibaud O., Clément J. M., Hofnung M. Structure of the malB region in Escherichia coli K12. III. Correlation of the genetic map with the restriction map. Mol Gen Genet. 1979 Jul 24;174(3):261–267. doi: 10.1007/BF00267798. [DOI] [PubMed] [Google Scholar]
- Raibaud O., Roa M., Braun-Breton C., Schwartz M. Structure of the malB region in Escherichia coli K12. I. Genetic map of the malK-lamB operon. Mol Gen Genet. 1979 Jul 24;174(3):241–248. doi: 10.1007/BF00267796. [DOI] [PubMed] [Google Scholar]
- Sancar A., Stachelek C., Konigsberg W., Rupp W. D. Sequences of the recA gene and protein. Proc Natl Acad Sci U S A. 1980 May;77(5):2611–2615. doi: 10.1073/pnas.77.5.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrenk W. J., Weisberg R. A. A simple method for making new transducing lines of coliphage lambda. Mol Gen Genet. 1975;137(2):101–107. doi: 10.1007/BF00341676. [DOI] [PubMed] [Google Scholar]
- Schumm J. W., Howe M. M. Mu-specific properties of lambda phages containing both ends of Mu depend on the relative orientation of Mu end DNA fragments. Virology. 1981 Oct 30;114(2):429–450. doi: 10.1016/0042-6822(81)90224-5. [DOI] [PubMed] [Google Scholar]
- Shimada K., Weisberg R. A., Gottesman M. E. Prophage lambda at unusual chromosomal locations. I. Location of the secondary attachment sites and the properties of the lysogens. J Mol Biol. 1972 Feb 14;63(3):483–503. doi: 10.1016/0022-2836(72)90443-3. [DOI] [PubMed] [Google Scholar]
- Shimada K., Weisberg R. A., Gottesman M. E. Prophage lambda at unusual chromosomal locations. II. Mutations induced by bacteriophage lambda in Escherichia coli K12. J Mol Biol. 1973 Oct 25;80(2):297–314. doi: 10.1016/0022-2836(73)90174-5. [DOI] [PubMed] [Google Scholar]
- Shuman H. A., Silhavy T. J., Beckwith J. R. Labeling of proteins with beta-galactosidase by gene fusion. Identification of a cytoplasmic membrane component of the Escherichia coli maltose transport system. J Biol Chem. 1980 Jan 10;255(1):168–174. [PubMed] [Google Scholar]
- Silhavy T. J., Benson S. A., Emr S. D. Mechanisms of protein localization. Microbiol Rev. 1983 Sep;47(3):313–344. doi: 10.1128/mr.47.3.313-344.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy T. J., Brickman E., Bassford P. J., Jr, Casadaban M. J., Shuman H. A., Schwartz V., Guarente L., Schwartz M., Beckwith J. R. Structure of the malB region in Escherichia coli K12. II. Genetic map of the malE,F,G operon. Mol Gen Genet. 1979 Jul 24;174(3):249–259. doi: 10.1007/BF00267797. [DOI] [PubMed] [Google Scholar]
- Soberon X., Covarrubias L., Bolivar F. Construction and characterization of new cloning vehicles. IV. Deletion derivatives of pBR322 and pBR325. Gene. 1980 May;9(3-4):287–305. doi: 10.1016/0378-1119(90)90328-o. [DOI] [PubMed] [Google Scholar]