Abstract
Studies of meiotic recombination have revealed an evolutionary paradox. Molecular and genetic analysis has shown that crossing over initiates at specific sites called hotspots, by a recombinational-repair mechanism in which the initiating hotspot is replaced by a copy of its homolog. We have used computer simulations of large populations to show that this mechanism causes active hotspot alleles to be rapidly replaced by inactive alleles, which arise by rare mutation and increase by recombination-associated conversion. Additional simulations solidified the paradox by showing that the known benefits of recombination appear inadequate to maintain its mechanism. Neither the benefits of accurate segregation nor those of recombining flanking genes were sufficient to preserve active alleles in the face of conversion. A partial resolution to this paradox was obtained by introducing into the model an additional, nonmeiotic function for the sites that initiate recombination, consistent with the observed association of hotspots with functional sites in chromatin. Provided selection for this function was sufficiently strong, active hotspots were able to persist in spite of frequent conversion to inactive alleles. However, this explanation is unsatisfactory for two reasons. First, it is unlikely to apply to obligately sexual species, because observed crossover frequencies imply maintenance of many hotspots per genome, and the viability selection needed to preserve these would drive the species to extinction. Second, it fails to explain why such a genetically costly mechanism of recombination has been maintained over evolutionary time. Thus the paradox persists and is likely to be resolved only by significant changes to the commonly accepted mechanism of crossing over.
Meiotic crossing over between homologous chromosomes plays two important roles in the genetic reshuffling caused by sexual reproduction: it creates new combinations of alleles within each chromosome, and it prevents nondisjunction during chromosome segregation (1). Crossovers are initiated primarily at specific sites called hotspots (2, 3). Most studies of the mechanism have been done in ascomycete fungi, where meiosis can be synchronously induced and all the meiotic products recovered in the ascus. Biased gene conversion is a typical consequence of recombination at hotspots (4–6). For example, a cross between active and inactive alleles of the ARG4 hotspot in the yeast Saccharomyces cerevisiae exhibits a 22-fold bias toward conversion of the active allele to its inactive homolog (6). Physical studies have shown that recombination events in S. cerevisiae are initiated by site-specific double-strand DNA breaks at hotspots, before the visible pairing of the chromosomes (7, 8). As illustrated in Fig. 1, these breaks are thought to be repaired by DNA synthesis that uses the strands of the homologous chromosomes as templates, with resolution of the repair intermediate frequently creating a crossover between the participating chromosomes (9, 10). This and related recombinational-repair mechanisms can account for both the preferential conversion of active hotspot alleles to their inactive homologs, seen in fungi, and the association of gene conversion with recombination hotspots, seen in many organisms (reviewed in ref. 2).
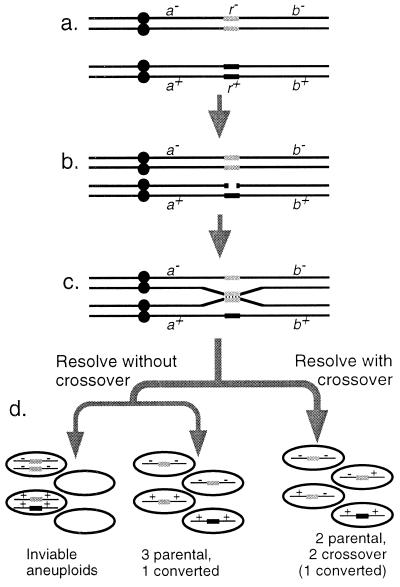
Figure 1.
Meiotic recombination in yeast and its consequences. (a) Two homologs of a single chromosome with a single recombination hotspot; r+ is an active hotspot allele (black), and r− is an inactive allele (gray). The chromosomes have replicated before meiosis; each black line represents a single chromatid. (b) One active hotspot allele undergoes a double-strand DNA break. (c) The broken chromatid undergoes recombinational repair, using a homologous chromatid as a template. In the process the broken hotspot is replaced with a copy of its homolog. (d) The recombining chromatids resolve, and completion of meiosis I and II produces four gametes: If resolution of the repair intermediate has produced a crossover, segregation is accurate and each gamete receives a single chromosome. If there has been no crossover, chromosomes may be distributed randomly at meiosis I, giving either four functional gametes or four aneuploid gametes.
But what accounts for the present abundance of hotspots despite their loss by conversion to inactive alleles? Even though inactive alleles may arise very rarely by mutation, they should take over the population by converting active alleles, unless their spread is opposed by selection, either for the benefits of crossing over or for other hotspot-associated cellular functions. To investigate the ability of these benefits to compensate for conversion, we have developed a computer model simulating a large population of diploid sexual organisms (diagrammed in Fig. 2).
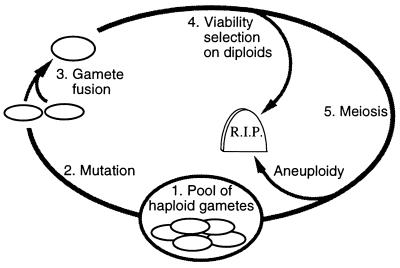
Figure 2.
The computer simulation model. A deterministic population was simulated using matlab (Student version 4a) on a Macintosh PowerPC 7200. Viability selection acts on diploids, mutation on gametes. The complete model is available by anonymous FTP at: ftp://ftp.zoology.ubc.ca/pub/redfield.
RESULTS
The Basic Model.
Each generation of the model begins (step 1) with a pool of haploid gametes, each containing as its genome a single chromosome with one recombination hotspot (initially all carry the active r+ allele). Mutant gametes containing an inactive hotspot allele (r−) arise (step 2) at frequency Mr (usually 10−8 per allele per generation). Diploids are formed by random fusion of gametes (step 3), and may be subject to viability selection (step 4) based on genotype at r and at two flanking loci a and b (described in more detail below). Individuals surviving selection undergo a conventional meiosis (step 5) with or without recombination, producing four gametes as illustrated in Fig. 1. At the start of this meiosis each homolog is represented by two sister chromatids. r+ alleles initiate recombinational repair with probability C. Sequential rounds of repair occur if two or more chromatids initiate, so that all are repaired. If the cell is heterozygous at r, repair converts initiating r+ chromatids to r−. Each repair event resolves as a crossover with probability X (usually X was set to 0.5). In this model, segregation is perfect if a crossover is present, and random otherwise (50% probability of four aneuploid gametes).
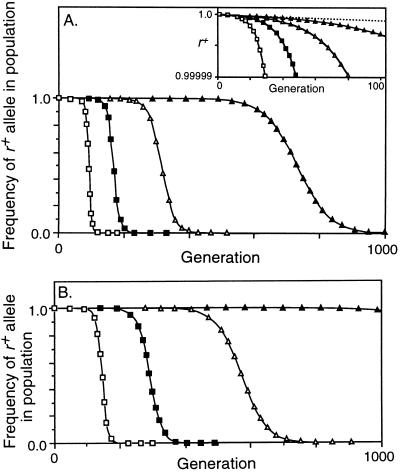
To first evaluate the power of the biased conversion of active hotspots, unopposed by any benefits, crossing over was eliminated by setting X = 0. Fig. 3A shows that r− alleles took over the population rapidly even when they arose very rarely (Mr = 10−8). The inset to Fig. 3A expands the scale during the apparent lag period to show that the rare r− alleles were increasing exponentially due to conversion; the dotted line shows the much slower change due to mutation alone. Changing C, the probability that active alleles initiate recombination, changed the rate of decrease of r+; the range of values of C shown in Fig. 3A corresponds to that characterized for naturally occurring hotspot alleles (11–16). Increasing or decreasing the mutation rate (to Mr = 10−6 or 10−10) changed only the delay until heterozygotes became common, by about 50 generations when C = 0.1 and by about 230 generations when C = 0.02 (not shown).
Figure 3.
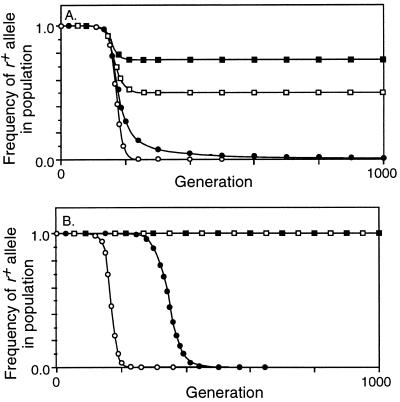
Decrease in frequency of the r+ allele due to conversion. Mr = 10−8, Mab = 0, no viability selection on a, b, or r. □, C = 0.2; ▪, C = 0.1; ▵, C = 0.05; ▴, C = 0.02. (A) Conversion not opposed by selection (X = 0). The inset is an enlargement of the first 100 generations, with the dotted line showing the effect of mutation alone. (B) Conversion opposed by fertility selection (X = 0.5). With C = 0.02 the frequency of the r+ allele had decreased to 0.01 at generation 1813.
This analysis confirmed that the conversion associated with the initiation of recombination can cause rapid extinction of active initiation sites. We next used the model to determine whether crossing over’s known benefits could successfully oppose this loss. We evaluated two well established benefits: prevention of aneuploidy and recombination between linked loci.
Segregation Benefits.
Meiotic crossing over has a well demonstrated role in the faithful segregation of homologous chromosomes (1, 17–19); homologs not connected by crossovers in meiotic prophase often fail to attach stably to the meiotic spindle, and consequently may be distributed randomly to the daughter cells. Thus by promoting crossovers r+ alleles will increase fertility, and this advantage might allow them to persist in spite of their loss by conversion in heterozygotes. However, this benefit is limited by two factors. First, not all break-and-repair events lead to chiasmata; simple isomerization models predict that one-half of conversion events will be accompanied by crossovers, but in S. cerevisiae and Neurospora crassa the ratio is only about one-third (20). Second, many species have backup systems that improve segregation in the absence of crossovers (19, 21).
In evaluating the segregation benefits of crossovers we deliberately exaggerated the benefit conferred by r+ alleles by (i) setting X = 0.5 so that half of all initiation events led to crossovers, with every crossover ensuring correct segregation, and (ii) having chromosomes disjoin randomly in the absence of a crossover. Although this benefit increased the fertility of r+ homozygotes from 0.5 to 0.55 when C = 0.05, and from 0.5 to 0.67 when C = 0.2, it did not prevent the invasion and ultimate fixation of r− alleles (Fig. 3B). As before, changing the mutation rate shifted the curves left or right without changing their shape. Simulations were also run with X = 1, so that every conversion event caused a crossover, but even this unrealistic condition did not prevent elimination of the r+ allele (not shown). The explanation may arise from the success of r− alleles in heterozygotes: when the r+ allele causes a crossover, the r− allele gets three-quarters of the benefits, and when the r+ allele causes conversion without crossover, the r− allele gets all the benefits.
Recombination Benefits.
To evaluate the benefits of the genetic recombination that crossing over can cause, two loci flanking r were introduced (a and b) into the model. Both were subject to viability selection and to deleterious mutation at rate Mab per gamete. It is well established that epistatic fitness interactions between genes can generate nonrandom associations between their alleles (linkage disequilibrium), and that a “modifier” gene that causes recombination between them can be selected because it breaks down these associations (22, 23). We tested fitness functions with no, moderate, and high negative epistasis [multiplicative, additive, and quadratic functions, respectively (24)] using selection coefficients that gave polymorphic equilibria at a and b, and an unnaturally high mutation rate (Mab = 0.1) chosen to exaggerate the benefit of recombination. Table 1 and Fig. 4 show results of these simulations.
Table 1.
Effects of genetic recombination
| Population/selection | Line | Fertility selection | Mr | Initial r+ | Equilibrium r+ | Equilibrium frequency of a+, b+ | Equilibrium viability | Equilibrium fertility |
|---|---|---|---|---|---|---|---|---|
| Asexual population | 1 | n.a. | n.a. | n.a. | n.a. | 0.3704 | 0.6561 | n.a. |
| Sexual/multiplicative selection | 2 | Off | 0 | 0 | 0 | 0.3704 | 0.6561 | n.a. |
| Wab = (1 − 0.15)i | 3 | Off | 0 | 1.0 | 1.0 | 0.3704 | 0.6561 | n.a. |
| 4 | On | 0 | 1.0 | 1.0 | 0.3704 | 0.6561 | 0.671 | |
| 5 | Off | 10−8 | 1.0 | 0 (155) | 0.3704 | 0.6561 | n.a. | |
| 6 | On | 10−8 | 1.0 | 0 (256) | 0.3704 | 0.6561 | 0.5 | |
| Sexual/additive selection | 7 | Off | 0 | 0 | 0 | 0.5198 | 0.6807 | n.a. |
| Wab = 1 − 0.15i | 8 | Off | 0 | 1.0 | 1.0 | 0.5300 | 0.6862 | n.a. |
| 9 | On | 0 | 1.0 | 1.0 | 0.5321 | 0.6873 | 0.6712 | |
| 10 | Off | 10−8 | 1.0 | 0 (155) | 0.5198 | 0.6807 | n.a. | |
| 11 | On | 10−8 | 1.0 | 0 (256) | 0.5198 | 0.6807 | 0.5 | |
| Sexual/quadratic selection | 12 | Off | 0 | 0 | 0 | 0.5184 | 0.7427 | n.a. |
| Wab = 1 − 0.02i − 0.04i2 | 13 | Off | 0 | 1.0 | 1.0 | 0.5463 | 0.7576 | n.a. |
| 14 | On | 1.0 | 1.0 | 0.5512 | 0.7601 | 0.6712 | ||
| 15 | Off | 10−8 | 1.0 | 0 (155) | 0.5184 | 0.7427 | n.a. | |
| 16 | On | 10−8 | 1.0 | 0 (256) | 0.5184 | 0.7427 | 0.5 |
In all simulations the initial frequency of a+ and b+ alleles was 1.0, and Mab = 0.1. Where fertility selection or conversion were acting, C = 0.2 and X = 0.5. The value in brackets is the number of generations taken for the r+ allele frequency to fall below 10−6. n.a., not applicable.
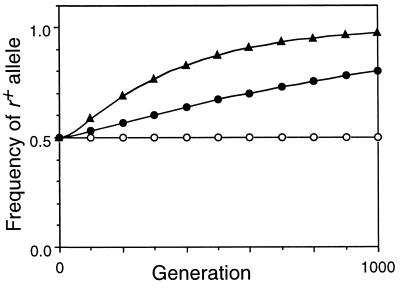
Figure 4.
Effect of selection against a− and b− mutations on frequency of the r+ allele. Mr = 0, C = 0.2, X = 0.5, Mab = 0.1, conversion at r and fertility selection inactivated. ○, multiplicative selection (Wab = 0.85i); •, additive selection (Wab = 1 − 0.15i); ▴, quadratic selection (Wab = 1 − 0.04i − 0.02i2).
The no-conversion control conditions in Table 1 (lines 2, 7, and 12) confirmed that, in the absence of crossing over, all functions gave equilibrium viabilities corresponding to those predicted analytically. For the multiplicative function this was (1 − Mab)4, the fraction of the population receiving no new mutations in each generation. This was identical to the viability of an asexual population and was not changed by recombination (compare lines 1, 2, and 3). With the epistatic selection functions, the equilibrium viabilities expected in the absence of crossovers at r were higher because of chromosome reassortment during meiosis, and were further increased by recombination at r (compare lines 7 and 8, and 12 and 13). [For the special cases in lines 7 and 12 the equations for the equilibria simplified to a four-dimensional system of cubics, which were solved explicitly using the symbolic package Maple.] Imposition of fertility selection further increased the viability slightly by eliminating some nonrecombinant gametes (lines 9 and 14). The effects of crossing over on viability were small, but Fig. 4 shows that in the absence of conversion they were sufficient to cause substantial increases in the frequency of r+ alleles.
However, when gene conversion was restored to the model, the benefit of recombination could not even slow the extinction of r+ alleles (compare lines 10 and 15 of Table 1 to line 5 and to Fig. 3A). When crossovers were assumed to increase fertility by ensuring correct segregation, active hotspot alleles were, as expected, lost more slowly, but again the benefit of recombination between a and b was too weak to influence the rate of loss (compare lines 11 and 16 to line 6 and to Fig. 3B).
Nonmeiotic Benefits.
The above results imply that meiotic recombination hotspots should be rapidly eliminated in spite of the benefits they confer in meiosis. Yet this has not occurred. This paradox might be resolved if these sites were found to be maintained by selection for some other cellular function. This possibility has been suggested by molecular studies, which have found that hotspots do not show any sequence consensus, but commonly coincide with transcription factor binding sites (13, 25), DNA replication origins (26), and other exposed regions of the chromosome (11, 27). Furthermore, genetic manipulations that alter chromatin structure or binding of transcription factors also alter hotspot activity (11, 28). Thus loci that act as hotspots in meiosis may have roles in cellular processes other than recombination (3), and selection for these other functions could balance loss of hotspots by conversion (6).
To evaluate this in the model, the viability of individuals homozygous for r− alleles was reduced by a selection coefficient Sr. Fig. 5 shows that this viability selection could balance or overcome loss of r+ by conversion, even in the absence of segregation and recombination benefits (X = 0; Mab = 0). When heterozygotes were assumed to be fully viable (Fig. 5A), strong selection caused the r+ and r− alleles to reach stable equilibria, although weaker selection only delayed extinction of r+. When r− and r+ were codominant (Fig. 5B), r− could invade to fixation when Sr was small, but was unable to invade when Sr was large; polymorphisms were not stable. This dependence on the mode and strength of selection is not surprising, as this version of the model is formally identical to a simple meiotic drive system countered by selection and gives identical results (29). As expected, restoring the segregation benefits (X = 0.5, Fig. 6) did decrease the amount of viability selection needed to give polymorphic equilibria and resistance to invasion. In contrast, the outcome was not changed by including the benefits of recombining a and b (data not shown), confirming the conclusion that recombination per se has very little effect on hotspot loss due to conversion.
Figure 5.
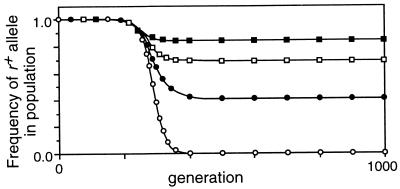
Effect of viability selection against r− mutations on frequency of the r+ allele. Mr = 10−8, C = 0.1, X = 0, Mab = 0, no viability selection on a or b. ○, Sr = 0; •, Sr = 0.1; □, Sr = 0.2; ▪, S4 = 0.4. (A) Wr++ = Wr+− = 1, Wr−− = 1 − Sr; (B) Wr++ = 1, Wr+− = 1 − 0.5Sr, Wr−− = 1 − Sr.
Figure 6.
Combined effects of fertility selection and viability selection against a−, b−, and r− mutations on frequency of the r+ allele. C = 0.1, Mr = 10−8, X = 0.5, Mab = 0, quadratic selection against a− and b−, (see legend to Fig. 4). ○, Sr = 0; •, Sr = 0.1; □, Sr = 0.2; ▪, Sr = 0.4. Wr++ = Wr+− = 1, Wr− = 1 − Sr.
The selection coefficients required to maintain an active allele at a single hotspot locus substantially reduced organismal fitness. The analysis in Table 2 shows that, if such viability selection were to act on the many loci needed to give experimentally observed crossover frequencies, the fitness cost would be intolerably high.
Table 2.
Analysis of hotspot loci maintained by viability selection
| Type of r locus | C | Sr | h | Frequency of r+ per locus at equilibrium | r loci needed/chromosome | Viability at equilibrium (1 locus) | Viability at equilibrium (10 chromosomes) |
|---|---|---|---|---|---|---|---|
| 1 | 0.01 | 0.0075 | 0 | 0.322 | 716 | 0.9966 | 2.57 × 10−11 |
| 2 | 0.1 | 0.280 | 824 | 0.9958 | 8.68 × 10−16 | ||
| 3 | 0.01 | 0 | 0.490 | 470 | 0.9974 | 4.85 × 10−6 | |
| 4 | 0.1 | 0.490 | 471 | 0.9969 | 4.46 × 10−7 | ||
| 5 | 0.02 | 0 | 0.744 | 310 | 0.9987 | 1.77 × 10−2 | |
| 6 | 0.1 | 0.807 | 285 | 0.9986 | 1.84 × 10−2 | ||
| 7 | 0.04 | 0 | 0.872 | 264 | 0.9993 | 0.175 | |
| 8 | 0.1 | 0.968 | 238 | 0.9997 | 0.497 | ||
| 9 | 0.1 | 0.075 | 0 | 0.236 | 99 | 0.9563 | 5.95 × 10−20 |
| 10 | 0.1 | 0.190 | 124 | 0.9484 | 3.07 × 10−29 | ||
| 11 | 0.1 | 0 | 0.411 | 57 | 0.9653 | 1.81 × 10−9 | |
| 12 | 0.1 | 0.406 | 58 | 0.9598 | 4.62 × 10−11 | ||
| 13 | 0.2 | 0 | 0.692 | 33 | 0.9810 | 1.78 × 10−3 | |
| 14 | 0.1 | 0.761 | 30 | 0.9813 | 3.47 × 10−3 | ||
| 15 | 0.4 | 0 | 0.842 | 27 | 0.9901 | 6.72 × 10−2 | |
| 16 | 0.1 | 0.955 | 24 | 0.9957 | 0.356 | ||
| All 16 types | Equal crossovers by each type of locus | 6.05 × 10−8 | |||||
| All 16 types | Equal numbers of each type of locus | 1.46 × 10−5 | |||||
This analysis assumed a haploid chromosome number of 10, and was done for each of 16 types of hotspot loci, differing in their assumed activities (C) and roles in viability (Sr and h). The first step was to use the above model to determine, for a single locus of each type, the equilibrium frequency of r+ and the consequent reduced viability. Segregation benefits were incorporated as in Fig. 3B, and the effects of recombination between a and b were eliminated by setting Mab = 0. The equilibrium frequency of r+ was then used to calculate the number of hotspot loci each chromosome required to ensure at least one crossover in 99% of meioses. With 10 chromosomes this ensured that less than 5% of gametes would be aneuploid due to lack of a crossover. Actual aneuploidy rates may be somewhat lower (30), so this analysis may underestimate the number of hotspots that must be maintained by selection. The calculated number of hotspots then was used to calculate the mean viability of the population at equilibrium, under the assumption that selection acted independently on each r locus.
The results, shown in the rightmost column of Table 2, imply that the cost of maintaining hotspots by viability selection would be intolerable. The magnitude depended on both the activity of the hotspot type and on its selective importance. However, even in the best case (type 8) more than half of the population died because of gene conversion at hotspot loci. Experimentally characterized genomes typically have heterogeneous hotspot loci; these are considered by the bottom two rows of Table 2. Here selection costs were evaluated assuming either that each of the 16 previously considered locus types contributed an equal number of crossovers, or that each type contributed an equal number of loci. In both cases the selection costs were extremely high.
DISCUSSION
Although the model neglects many factors seen in real meioses, the following points suggest that none of these factors is likely to mitigate the extinction of active hotspots.
Mutation.
Because extinction is driven by conversion rather than mutation, the outcomes are extremely insensitive to changes in the rate of mutation at r. Back mutation from r− to r+ is unlikely to be significant. Simple algebra shows that the equilibrium frequency of the r+ allele can be well approximated by the back mutation rate divided by twice the r+ alleles’ frequency of initiation (C). Thus, unless the back mutation rate is extraordinarily high, new r+ alleles will normally occur in r+/r− heterozygotes, where they will reconvert themselves to r− the first time they act. The model does not explicitly consider mutation to alleles of r having low initiation activity (rather than no activity), but the outcome should depend only on the difference in activity between the competing alleles. The genes a and b flanking r are assumed to mutate at an unrealistically high rate, but reducing this would only decrease the very weak benefits of recombination.
Selection and Recombination.
The strong epistatic selection acting on the flanking genes, like the high mutation rate, serves to maximize the recombination benefits opposing loss of hotspots. The benefits cannot be increased further without causing most of the population to be killed by selection in each generation. For the same reason, introducing additional flanking genes subject to epistatic selection would require weakening selection and mutation pressures on each gene, and thus will not significantly increase the benefits of recombination between the loci. The model does not evaluate the genetic benefits of recombination in small populations (Muller’s Ratchet; ref. 31), nor the advantage of recombination in the presence of rapidly evolving pathogens or parasites (32). However, the effects of these benefits are unlikely to be substantially greater than those of recombining deleterious mutations, which were tested and found to be extremely weak relative to the cost of conversion.
Genome Structure.
Increasing the number of chromosomes per haploid set, or the number of hotspots per chromosome, will not strengthen the segregation or recombination benefits. Nonhomologous chromosomes segregate independently, so the presence of additional chromosomes would not improve each chromosome’s ability to segregate correctly from its homolog and would expose it to the additional risk of segregating into a gamete that had become aneuploid for a different chromosome. Chromosomes typically have multiple hotspots and undergo at least one and no more than several crossovers in each meiosis, with interference between hotspots limiting the number of crossovers. But interference acts only to limit crossing over, not conversion (33), so introducing multiple hotspots per chromosome into the model would reduce each hotspot’s contribution to accurate segregation and to recombination without reducing its risk of conversion.
Thus the model appears to be robust; the conclusion that the sites thought to initiate crossing over cannot be maintained by the benefits of the events they cause is not an artefact caused by simplifying assumptions.
Is this self-destructive recombination mechanism the only possible way to carry out meiotic recombination? The need to correctly pair many sites distributed throughout a large genome may preclude a simple cut-and-ligate mechanism like those used by most site-specific recombination systems, unless a different recognition sequence were used for each site, or homology reliably established by paranemic pairing of flanking sequences before the actual initiation of recombination (34). A mechanism where the recognition site caused its homolog to be cut (cutting-in-trans) would prevent loss of active hotspots by conversion, but, because this would reverse the direction of the conversion bias, could cause hyperactive hotspots to overrun the genome. However even with cutting-in-cis there is no evident need for conversion at the initiation site, as the free ends required for an efficient homology search by plectonemic pairing could in principle be generated by a simple nick or break without accompanying strand degradation.
The existence of self-converting hotspots at functional sites could be a consequence of using a recombinational-repair mechanism to initiate meiotic crossovers. Recombinational repair is already an essential component of any cell’s DNA repair tool kit and appears to have been adapted for meiotic crossing over in yeast by production of a meiosis-specific endonuclease. Such a nuclease might be constrained by chromatin structure to act primarily at relatively exposed DNA, thus causing crossing over to initiate preferentially at active regions of the chromosome. Those sites whose roles in viability were weaker than their frequency of conversion would then be eliminated by the associated conversion to inactive alleles. This explanation makes the testable prediction that the only hotspots to persist will be under viability selection strong enough to compensate for their loss by conversion.
McKee (35) has suggested that initiation of crossing over at promoters is an adaptation that targets recombination to transcriptionally active and hence mutation-prone regions of the genome, especially those containing promoter-up mutations. However, this neglects the high cost of the selection needed to maintain normal promoters and also requires that mutation rates be higher than conversion frequencies, when in reality they are at least several orders of magnitude lower (36).
Consideration of direct viability selection on hotspots does not fully resolve the paradox of hotspot persistence, because the population’s reproductive capacity may be drastically reduced by the partial or complete loss of alleles for many important functions. The computer model we have presented readily incorporates this selection, because it considers only a single hotspot in an organism with unlimited reproductive capacity. However, providing at least one crossover per chromosome would require tens or hundreds of moderately active hotspots, an estimate consistent with the distribution of recombination sites and the distributions and frequencies of meiotic double-strand breaks (11).
The cost of this viability selection is very sensitive to the selection parameters acting on the individual loci. At one extreme, r+ alleles whose selection coefficients are well below the frequency with which the allele initiates recombination will be eliminated by conversion, and their contributions to viability permanently lost. At the other extreme, loci whose r− alleles are quickly eliminated by strong codominant selection will retain r+ alleles at little cost. However, if, as suggested above, viability selection is due to various nonmeiotic functions causing an open chromatin structure, many hotspot loci will fall between these extremes, and selection on these will maintain polymorphisms like those analyzed in Table 2. The cost this selection imposes need not be a serious problem for yeast and other facultatively sexual organisms, where it could be spread over the many asexual generations occurring between rare sexual events. However, such strong selection could easily drive obligately sexual species to extinction.
In summary, the paradox remains unresolved at two levels. First, the mechanism of meiotic recombination, including initiation at hotspots, presumably exists to cause crossing over, but our analysis has failed to identify benefits of crossing over strong enough to maintain hotspots in the face of their loss by conversion. Second, if initiation at hotspots is an accidental aspect of recombination, it is an extremely expensive one, because it causes functional sites such as active promoters to be replaced by inactive alleles. We are left with the possibility that the role of initiation hotspots in meiotic crossing over has been misinterpreted.
Acknowledgments
We thank Jim Haber, Sally Otto, Frank Stahl, and Gerry Smith for helpful discussions, and Marc Feldman, Michael Lichten, Wen-Hsiung Li, and Ken Hillers for comments on the manuscript. R.J.R. is a Scholar of the Evolutionary Biology Program of the Canadian Institute for Advanced Research. This work was supported by an operating grant from the Natural Sciences and Engineering Research Council of Canada.
References
- 1.Murray A W, Szostak J W. Annu Rev Cell Biol. 1985;1:289–315. doi: 10.1146/annurev.cb.01.110185.001445. [DOI] [PubMed] [Google Scholar]
- 2.Nicolas A, Petes T D. Experientia. 1994;50:242–252. doi: 10.1007/BF01924007. [DOI] [PubMed] [Google Scholar]
- 3.Smith G R. Experientia. 1994;50:234–238. doi: 10.1007/BF01924006. [DOI] [PubMed] [Google Scholar]
- 4.Grimm C, Schaer P, Munz P, Kohli J. Mol Cell Biol. 1991;11:289–298. doi: 10.1128/mcb.11.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Catcheside D G. Aust J Biol Sci. 1975;28:213–225. doi: 10.1071/bi9750213. [DOI] [PubMed] [Google Scholar]
- 6.Nicolas A, Treco D, Schultes N P, Szostak J W. Nature (London) 1989;338:35–39. doi: 10.1038/338035a0. [DOI] [PubMed] [Google Scholar]
- 7.Padmore R, Cao L, Kleckner N. Cell. 1991;66:1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- 8.Goyon C, Lichten M. Mol Cell Biol. 1993;13:373–382. doi: 10.1128/mcb.13.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szostak J W, Orr-Weaver T L, Rotstein R J, Stahl F W. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 10.Schwacha A, Kleckner N. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- 11.Wu T C, Lichten M. Science. 1994;263:515–518. doi: 10.1126/science.8290959. [DOI] [PubMed] [Google Scholar]
- 12.Zahn-Zabal M, Lehmann E, Kohli J. Genetics. 1995;140:469–478. doi: 10.1093/genetics/140.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shiroishi T, Koide T, Yoshino M, Sagai T, Moriwaki K. Adv Biophys. 1995;31:119–132. doi: 10.1016/0065-227x(95)99387-5. [DOI] [PubMed] [Google Scholar]
- 14.Schultes N P, Szostak J W. Genetics. 1990;126:813–822. doi: 10.1093/genetics/126.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patterson G I, Kubo K M, Shroyer T, Chandler V L. Genetics. 1995;140:1389–1406. doi: 10.1093/genetics/140.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Catcheside D E A. Aust J Biol Sci. 1970;23:855–865. doi: 10.1071/bi9700855. [DOI] [PubMed] [Google Scholar]
- 17.Hassold T J, Sherman S L, Pettay D, Page D C, Jacobs P, A. Am J Hum Genet. 1991;49:253–260. [PMC free article] [PubMed] [Google Scholar]
- 18.Goldway M, Sherman A, Zenvirth D, Arbel T, Simchen G. Genetics. 1993;133:159–169. doi: 10.1093/genetics/133.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hawley R S, McKim K S, Arbel T. Annu Rev Genet. 1993;27:281–317. doi: 10.1146/annurev.ge.27.120193.001433. [DOI] [PubMed] [Google Scholar]
- 20.Foss E, Lande R, Stahl F W, Steinberg C M. Genetics. 1993;133:681–691. doi: 10.1093/genetics/133.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loidl J, Scherthan H, Kaback D B. Proc Natl Acad Sci USA. 1994;91:331–334. doi: 10.1073/pnas.91.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barton N H. Genet Res. 1995;65:123–145. doi: 10.1017/s0016672300033140. [DOI] [PubMed] [Google Scholar]
- 23.Feldman M W, Christiansen F B, Brooks L D. Proc Natl Acad Sci USA. 1980;77:4838–4841. doi: 10.1073/pnas.77.8.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crow J F. In: Mathematical Topics in Population Genetics. Kojima K, editor. Berlin: Springer; 1970. pp. 128–177. [Google Scholar]
- 25.White M A, Dominska M, Petes T D. Proc Natl Acad Sci USA. 1993;90:6621–6625. doi: 10.1073/pnas.90.14.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rattray A J, Symington L S. Genetics. 1993;134:175–188. doi: 10.1093/genetics/134.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohta K, Shibata T, Nicolas A. EMBO J. 1994;13:5754–5763. doi: 10.1002/j.1460-2075.1994.tb06913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu T C, Lichten M. Genetics. 1995;140:55–66. doi: 10.1093/genetics/140.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hartl D L, Clark A G. Principles of Population Genetics. Sunderland, MA: Sinauer; 1989. [Google Scholar]
- 30.Griffiths A J, Miller J H, Suzuki D T, Lewontin R C, Gelbart W M. An Introduction to Genetic Analysis. 6th Ed. New York: Freeman; 1996. [Google Scholar]
- 31.Muller H J. Mutat Res. 1964;1:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 32.Ebert D, Hamilton W D. Trends Ecol Evol. 1996;11:79–82. doi: 10.1016/0169-5347(96)81047-0. [DOI] [PubMed] [Google Scholar]
- 33.Mortimer R K, Fogel S. In: Mechanisms in Recombination. Grell R F, editor. New York: Plenum; 1974. pp. 263–275. [Google Scholar]
- 34.Weiner B M, Kleckner N. Cell. 1994;77:977–991. doi: 10.1016/0092-8674(94)90438-3. [DOI] [PubMed] [Google Scholar]
- 35.McKee B D. BioEssays. 1996;18:411–419. doi: 10.1002/bies.950180511. [DOI] [PubMed] [Google Scholar]
- 36.Datta A, Jinks-Robertson S. Science. 1995;268:1616–1619. doi: 10.1126/science.7777859. [DOI] [PubMed] [Google Scholar]