Abstract
The lineage relationships between murine CD8+ T cells with different cytokine profiles were investigated by paired-daughter analysis in the presence and absence of the type 2 cytokine-inducing stimulus, interleukin 4 (IL-4). Single CD8+ CD44low lymph node T cells were activated to divide at high frequency with IL-2 and immobilized antibodies to CD3, CD8, and LFA-1. When these parent cells were subcloned by transferring their daughter or granddaughter cells into secondary cultures with or without IL-4, the subclones expressed diverse combinations of the mRNAs for the type 1 cytokines, interferon γ (IFN-γ), and IL-2, and the type 2 cytokines, IL-4, IL-5, IL-6, and IL-10. Frequencies of subclones that expressed IL-4, IL-6, and, to a lesser extent, IL-2, IL-5, and IL-10 were higher among those grown with IL-4, but a significant proportion of those grown without exogenous IL-4 also expressed one or more type 2 cytokines. Subclones within 89% of families displayed different cytokine profiles, indicating that their parent cells were multipotential for this function. Because 98% of parent cells yielded subclones that produced type 1 cytokines and 77% yielded type 2 cytokine producers, we conclude that type 1 and type 2 cytokine-producing CD8+ T cells can be derived from a common precursor. Similar analyses performed by subcloning after ≥7 or ≥13 cell divisions without IL-4 showed that many CD8+ T cells retained the potential to shift toward a type 2 cytokine profile in response to IL-4, even after prolonged expansion under conditions that favored type 1 cytokine expression. CD8+ T cells that express type 1 and/or type 2 cytokines therefore are derived from the same peripheral T cell lineage whose multipotentiality can persist through many cell divisions.
Activated murine CD4+ T cells can synthesize many different combinations of cytokines. Both in vitro and in vivo, CD4+ T cell populations can be polarized toward the preferential synthesis of type 1 cytokines, such as interleukin 2 (IL-2) and interferon γ (IFN-γ), or type 2 cytokines, such as IL-4, IL-5, IL-6, and IL-10 (1, 2). Because clones with different profiles can be derived from a single CD4+ T cell (3, 4), the acquisition of a certain cytokine profile apparently occurs by differentiation of uncommitted peripheral T cells after activation in the presence of polarizing stimuli (1, 2). The best characterized of these stimuli are IL-12 for the type 1 pathway and IL-4 for the type 2 pathway.
The situation is less clear for murine CD8+ T cells. Recent work in several laboratories has shown that normal CD8+ T cell populations can synthesize type 2 cytokines when cultured with IL-4 (5–8). However, in most immune reactions where CD8+ T cells are activated, such as alloreactive and antiviral responses, the responding CD8+ cells and clones derived from them preferentially express IFN-γ (9–11); although examples of in vivo-activated CD8+ type 2 cytokine producers have been reported (12–15), these remain the exception. Even CD8+ T cells from mice continuously exposed to IL-4 by transgenic overexpression (16) or undergoing a type 2-polarized CD4+ T cell response (ref. 17; A. G. Doyle and A.K., unpublished observations) preferentially display a type 1 cytokine profile. This discrepancy between the in vitro potential of CD8+ T cell populations and their usual behavior in vivo raises the possibility that type 2 cytokine-producing CD8+ T cells are derived from a distinct and minor peripheral T cell lineage, which can be expanded in response to IL-4 in vitro, but is not responsible for the dominant CD8+ T cell response to common class I-dependent immunogens in vivo. Previous studies have not distinguished whether CD8+ type 2 cytokine producers arise from the same precursors as the conventional type 1 cytokine producers.
Here we describe the resolution of this issue using paired-daughter analysis. This experimental approach has been used by others, for example, to determine lineage relationships between blood cell types and to distinguish differentiative and proliferative actions of cytokines (18, 19). We have used a culture system in which the combination of immobilized antibodies to CD3ɛ, CD8, and LFA-1 with IL-2 stimulates clone formation by up to 75% of single CD8+ T cells in the absence of accessory cells (20, 21). These conditions induce synthesis of IL-2, IL-3, IFN-γ, granulocyte-macrophage colony stimulating factor, and tumor necrosis factor-α mRNA and/or protein without detectable IL-4 or IL-6 in bulk and clonal cultures (20). Inclusion of IL-4 induces the additional production of IL-4, IL-5, IL-6, and IL-10 mRNA and protein in bulk cultures (unpublished observations). The high cloning efficiency, IL-4 responsiveness, and accessory cell independence of this system enabled its use to assess whether paired progeny of individual CD8+ T cells could form clones with different cytokine profiles in the presence and absence of IL-4, or whether IL-4 selectively expanded a distinct subset of CD8+ T cells. We show here that most clonogenic CD8+ T cells are multipotential in their cytokine profiles for two or, in some cases, many divisions after primary activation, and that type 2 cytokine-producing CD8+ cells do not represent a distinct lineage.
MATERIALS AND METHODS
T Cell Preparation.
Specific pathogen-free female C57BL/6 mice were obtained from the Animal Resources Centre (Western Australia) and used at 6–8 weeks of age. Cell suspensions from brachial, axillary, inguinal, and para-aortic lymph nodes were stained with fluoresceinated anti-CD8 (53.6; Becton Dickinson) and biotinylated anti-CD44 (IM7; PharMingen) mAb followed by phycoerythrin-conjugated streptavidin (Caltag Laboratories, South San Francisco, CA) then resuspended with propidium iodide. Using a fluorescence-activated cell sorter (FACS Vantage with lysis ii software; Becton Dickinson), single viable cells selected for propidium iodide exclusion, forward and side scatter properties of small lymphocytes, expression of CD8 and low expression of CD44 (lowest 30% of distribution) were deposited into culture wells using an automated cell deposition unit attached to the FACS.
T Cell Cloning and Subcloning.
All cultures were performed in supplemented DMEM (21) with 12.5% fetal calf serum in the wells of mAb-coated plates. For cultures of 2–5 days, Terasaki microwells (Nunc) were incubated with purified mAb to CD3ɛ (145–2C11; 10 μg/ml), CD8 (53.6; 10 μg/ml), and LFA-1 (I21/7.7; 5 μg/ml), washed and the diluent replaced with 15 μl of medium containing 600 IU/ml of recombinant human IL-2 (Cetus) (20). Where clones were to be expanded for 7 days, single CD8+ CD44low T cells were deposited into flat-bottomed 96-well plates coated with mAb as above and containing 200 μl of medium and IL-2. Cultures were checked microscopically for viable cells, and, where the parent cells had divided one or more times, individual cells were transferred by micromanipulation into new Terasaki wells coated with the same mAb as above; at least one cell was cultured with IL-2 as above and one with IL-2 plus 100 units/ml of IL-4 [supernatant of Sf9 cells infected with murine IL-4 cDNA-containing baculovirus (22), depleted of IL-4 cDNA and RNA by ultrafiltration]. One unit/ml of IL-4 activity was defined as the amount stimulating half-maximal proliferation of the IL-4-dependent cell line CT.4S (22). After 3–5 days’ incubation, secondary cultures were checked microscopically for the presence of subclones and their size counted.
RNA Extraction and Cytokine mRNA Analysis by Reverse Transcription–PCR (RT-PCR).
Subclones were lysed in situ with guanidine thiocyanate (20) or Nonidet P-40 (23) then transferred to microfuge tubes. Subsequent steps in RNA extraction, reverse transcription, and cDNA amplification by two nested rounds of 35-cycle PCR were performed as described (20, 24, 25) with the following external (ex) and internal (in) intron-spanning primer pairs (5′ then 3′): β-actin, ex GACATGGAGAAGATCTGGCA, GGTCTTTACGGATGTCAACG, in CCCAGATCATGTTTGAGACCTTC, GCTCGTTGCCAATAGTGATGA; CD3ɛ, ex TGCGTCCGCCATCTTGGTAGA, CGCTCCTTGTTTTGCCCTCTG, in CTGAGAGGATGCGGTGGAACA, GACCATCAGCAAGCCCAGAGT; IFN-γ, ex CATGAAAATCCTGCAGAGCC, GGACAATCTCTTCCCCACCC, in CCTCAGACTCTTTGAAGTCT, CAGCGACTCCTTTTCCGCTT; IL-2, ex CAGCTCGCATCCTGTGTCAC, AAGGCTATCCATCTCCTCAG, in GTGCTCCTTGTCAACAGCGC, AGAACATGCCGCAGAGGTCCA; IL-4, ex TCTTTCTCGAATGTACCAGG, CATGGTGGCTCAGTACTACG, in CACTTGAGAGAGATCATCGG, GGCTTTCCAGGAAGTCTTTCA; IL-5, ex TTGACAAGCAATGAGACGAT, GGCTACATTACCAGTTTGAG, in TAATAAAGAAATACATTGACCGCC, ACACTTTGCATATATGGACATAGAT; IL-6, ex TGCTGGTGACAACCACGGCC, GTACTCCAGAAGACCAGAGG, in GAGGATACCACTCCCAACAG, CCAGTTTGGTAGCATCCATCA; and IL-10, ex CCAAAGCCACAAAGCAGCCT, GCTCTGTCTAGGTCCTG, in AGAGAGCTCCATCATGCCTG, CTCAATACACACTGCAGGTG.
Amplifications were performed in a single reaction for β-actin, IFN-γ, and IL-4, and for β-actin, IL-2, and IL-6. IL-5, IL-10, and CD3ɛ were amplified in separate reactions. PCR products were separated by gel electrophoresis, visualized with ethidium bromide, and identified by Southern hybridization (20). Cytokine and CD3ɛ PCR products of correct size were not detected if reverse transcription was omitted. All PCR runs included a titration of cloned β-actin and cytokine cDNAs to monitor cDNA sensitivity (at least 10−16 g) and at least 10 negative control samples to which cDNA was not added, as shown elsewhere (25). No PCR products were detected in any negative control samples. IL-4 PCR products were not detected when the filtered rIL-4 source was used as a template with or without reverse transcription in amounts up to 100-fold higher than those added to culture. The frequency of successful RNA extraction from small cell numbers was improved by use of Nonidet P-40 lysis (experiments 3 and 4 in Table 1) instead of guanidine thiocyanate lysis and phenol-chloroform extraction (experiments 1 and 2). Because genomic DNA was not removed by the Nonidet P-40 method and contained pseudogenes that could yield a β-actin PCR product of the same size as that from mRNA, samples obtained by this method were assayed for CD3ɛ mRNA; 100% yielded a CD3ɛ PCR product of the correct size to be encoded by mRNA.
Table 1.
Efficiencies of CD8+ CD44low LN T cell cloning and cytokine mRNA detection among subclones
| Parameter | Experiment 1 | Experiment 2 | Experiment 3 | Experiment 4 | ||||
|---|---|---|---|---|---|---|---|---|
| Primary cultures | ||||||||
| No. parent cells cultured | 480 | 480 | 180 | 300 | ||||
| % cultures with ≥1 cell at day 2 | 72% | 75% | 59% | 58% | ||||
| % cultures with ≥2 cells at day 2 | 69% | 62% | 46% | 52% | ||||
| Secondary cultures | IL-2 | IL-2 + IL-4 | IL-2 | IL-2 + IL-4 | IL-2 | IL-2 + IL-4 | IL-2 | IL-2 + IL-4 |
| No. cells transferred on day 2 | 78 | 75 | 58 | 56 | 68 | 60 | 58 | 51 |
| % cells that recloned by day 5 | 83% | 75% | 95% | 88% | 85% | 75% | 88% | 84% |
| Mean subclone size on day 5 | 52 ± 38 | 38 ± 32 | 66 ± 49 | 43 ± 40 | 106 ± 70 | 86 ± 69 | 130 ± 63 | 106 ± 54 |
| No. subclones assayed by RT-PCR | 55 (38) | 54 (38) | 44 (37) | 44 (37) | 42 (38) | 44 (38) | 30 (30) | 30 (30) |
| No. β-actin/CD3ɛ+ subclones | 37 (26) | 35 (26) | 25 (25) | 20 (18) | 42 (38) | 44 (38) | 30 (30) | 30 (30) |
| No. informative parent cells | 20 | 13 | 38 | 30 | ||||
Numbers in parentheses indicate the number of parent cells from which the assayed subclones were derived.
RESULTS
Multipotentiality of CD8+ T Cells Assessed by Paired-Daughter Analysis After One or Two Cell Divisions.
Single CD8+ T cells were enriched for naive cells based on small size and low CD44 expression (26, 27) and cultured in anti-receptor antibody-coated wells with IL-2 for 40–48 hr. At this stage, an average of 66% of cells had survived, and, of these, 87% had divided at least once (Table 1). Individual daughter or granddaughter cells from clones of 2–4 cells were transferred into new anti-receptor antibody-coated wells containing IL-2 or IL-2 + IL-4. Up to 95% of transferred cells recloned, suggesting that T cell receptor ligation did not cause activation-induced death at this stage. Both recloning efficiencies and subclone size were ≈2-fold higher in the presence of anti-receptor antibodies and IL-2 ± IL-4 than in the cytokines alone (data not shown). After 3–5 days’ incubation of secondary cultures, RNA was extracted from subclones in those families in which cells recloned successfully in both IL-2 and IL-2 + IL-4. The presence of β-actin, CD3ɛ, and cytokine mRNAs was assayed by RT-PCR and Southern hybridization using conditions shown to detect cytokine transcripts in single activated T cells (24, 25). Previous studies showed that immobilized anti-receptor antibodies remain stimulatory for at least 7 days at 37°C and that cytokine expression continues in bulk and clonal T cell cultures over the same period (20, 21). Two independent sets of PCR amplifications from the 109 cDNA samples in experiment 1 yielded results that were 90% identical; reproducibility was 100% for control amplifications performed in all experiments from cDNA derived from 5 × 104 CD8+ cells activated in the presence of IL-4 (data not shown).
In total, 101 parent cells yielded β-actin/CD3ɛ+ subclones in both the presence and absence of IL-4 and therefore could be used to assess whether different subclones of a single cell displayed different cytokine profiles. Taking into account frequencies of survival in primary culture (weighted mean 62%; Table 1), recloning of sibling pairs in both IL-2 and IL-2 + IL-4 (69%), and PCR product detection in subclone pairs (87%), these informative cells represented 37% of the starting CD8+ CD44low population.
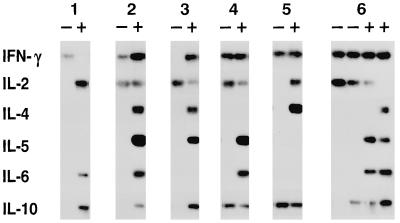
Fig. 1 shows data for six families of subclones derived after one or two divisions of the parent cell. Within each family, the subclones differed in the cytokine combinations expressed, indicating that the parent cell had not been committed to expression of a single combination and was therefore multipotential. In total, 50% of the 101 informative parent cells were multipotential based on expression of the representative type 2 cytokine, IL-4, and 89% were multipotential when all six cytokines were considered (Table 2). By χ2 analysis, these frequency estimates were statistically independent of the number of cell divisions before transfer. Pairwise tests of association did not detect any statistical relationship between cytokine profiles within subclone families. Multipotentiality also was detected when granddaughters were cultured in identical conditions (e.g., family 6 in Fig. 1): the members of 5 of 12 pairs grown in IL-2 and 12 of 15 pairs grown in IL-2 + IL-4 displayed different cytokine profiles.
Figure 1.
Paired-daughter analysis of the cytokine secretion potential of individual CD8+ T cells. Southern hybridization of RT-PCR products is shown for six families of subclones derived from two daughters (families 1–5) or four granddaughters (family 6) cultured in the absence (−) or presence (+) of IL-4 (from experiment 3 in Table 1).
Table 2.
Frequencies of multipotential cells detected among CD8+ CD44low T cells
| Cytokines considered | No. divisions before cell transfer | No. parent cells assayed | No. multipotential parent cells |
|---|---|---|---|
| IL-4 | 1 | 70 | 32 (46%) |
| 2 | 31 | 19 (61%) | |
| Total 101 | 51 (50%) | ||
| 6 cytokines | 1 | 70 | 60 (86%) |
| 2 | 31 | 30 (97%) | |
| Total 101 | 90 (89%) |
Parent cells were defined as multipotential if two of their subclones expressed different cytokine combinations. Frequencies were estimated considering expression of IL-4 alone, or all six assayed cytokines (IFN-γ, IL-2, IL-4, IL-5, IL-6, and IL-10), and for parent cells whose progeny were separated after either one or two divisions.
Effect of IL-4 on Clonal Cytokine Profiles.
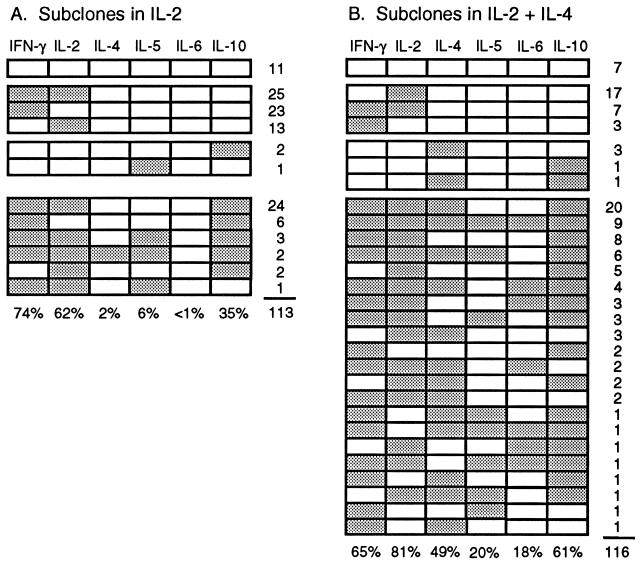
Fig. 2 summarizes the cytokine profiles of all the assayed subclones derived from the 101 informative parent cells in the presence or absence of IL-4. Whereas some subclones in both groups expressed the canonical type 1 cytokine profile (IFN-γ and IL-2 alone), none expressed the complete type 2 profile (IL-4, IL-5, IL-6 and IL-10 alone). Surprisingly, 36% of the subclones grown in IL-2 expressed one or more of the type 2 cytokines, IL-4, IL-5, and most frequently IL-10. Culture in IL-4 did, however, increase the frequency of type 2 cytokine producers to 72%. This effect was most marked for IL-4 itself (27-fold) and IL-6 (>20-fold) and more modest for IL-5 (3.2-fold) and IL-10 (1.8-fold). These changes were accompanied by a minor reduction in the frequency of IFN-γ producers (P > 0.05) and a significant increase in the frequency of IL-2 producers (P < 0.05).
Figure 2.
Cytokine mRNA expression patterns displayed by 229 subclones derived from 101 families of daughters and granddaughters cultured in the absence (A) or presence (B) of IL-4. Detection of a cytokine PCR product is indicated by shading of the boxes. Expression patterns are displayed from top to bottom in the groups: no cytokines, type 1 cytokines only, type 2 cytokines only, and mixed type 1 and type 2 cytokines. The number of subclones with each pattern is indicated at the right. The frequency of expression of each cytokine is indicated at the bottom.
Almost all parent CD8+ cells (98%) gave rise to one or more subclones that expressed a type 1 cytokine (IFN-γ and/or IL-2) and the majority (77%) gave rise to one or more subclones that expressed a type 2 cytokine. Even when IL-10 was excluded, most parent cells (62%) yielded a subclone that produced at least one of the other type 2 cytokines.
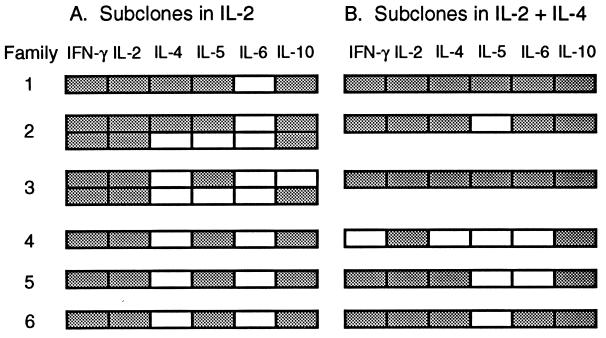
The finding that some subclones in Fig. 2A expressed IL-4 and/or IL-5 in the absence of in vitro exposure to exogenous IL-4 raises the possibility that their parent cells had undergone commitment during exposure to IL-4 in vivo. Fig. 3 shows that the cytokine profiles of these subclones differed from those of their siblings, arguing against precommitment of their parent cells to a single profile. Similarly heterogeneous patterns were observed within families of subclones that expressed IL-10 mRNA without in vitro exposure to exogenous IL-4 (data not shown).
Figure 3.
Cytokine mRNA expression patterns within families in which subclones produced IL-4 and/or IL-5 in the absence of exogenous IL-4. The cytokine profiles of the six subclones from Fig. 2A that produced IL-4 and/or IL-5 are aligned with the profiles of their siblings grown in either IL-2 (A) or IL-2 + IL-4 (B). Detection of a cytokine PCR product is indicated by shading of the boxes.
Multipotentiality of CD8+ T Cells Assessed after Extended Clonal Expansion.
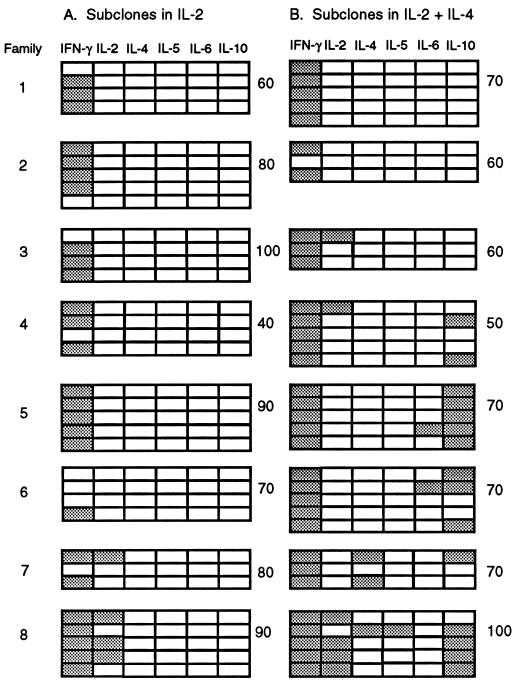
To determine whether the ability to respond to IL-4 by synthesizing type 2 cytokines was retained with longer clonal expansion, 12 primary clones were grown to sizes of 200–300 cells (≥7 divisions) over 5 days and, in another experiment, five primary clones were grown to sizes of 0.9–4.6 × 104 cells (≥13–15 divisions) over 7 days without added IL-4 before transfer of individual cells into secondary cultures with and without IL-4. Screening for IFN-γ and IL-4 expression by β-actin/CD3ɛ+ subclones showed that 2 of the 12 parent cells in the first experiment and 2 of the 5 parent cells (primary clones of 1.9 × 104 and 4.1 × 104 cells) in the second experiment yielded one or more IL-4-producing subclones when grown in IL-4; no subclones grown without exogenous IL-4 expressed this cytokine. Eight of the subclone families from the first experiment were assayed for expression of additional cytokines (Fig. 4). Five families included subclones that expressed one or more type 2 cytokines in the presence of IL-4 (70% of subclones in 63% of families); no subclones grown without IL-4 expressed these cytokines. We conclude that some cells retained the ability to respond to IL-4 by expressing type 2 cytokines despite prolonged clonal expansion in IL-2 alone.
Figure 4.
Cytokine mRNA expression patterns within families of subclones derived after ≥7 cell divisions. Primary clones were grown for 5 days with anti-receptor antibodies and IL-2 and those of 200–300 cells identified microscopically. Twenty individual cells from each clone were transferred into secondary cultures, 10 with IL-2 (A) and 10 with IL-2 + IL-4 (B). After 3 days, RNA was extracted from the five largest subclones of each group (mean 37 ± 31, range 2–130 cells) and assayed for β-actin and cytokine mRNAs. Results are shown for all β-actin+ subclones in eight families. Detection of a cytokine PCR product is indicated by shading of the boxes. Subcloning efficiencies (%) for each group of 10 transferred cells are shown at the right.
DISCUSSION
Here we show that activated CD8+ T cells with different cytokine profiles can develop from a common post-thymic precursor of naive phenotype. The finding that most lymph node CD8+ T cells analyzed gave rise to clones expressing one or more type 2 cytokines indicates that CD8+ T cells with this profile do not comprise a minor T cell lineage distinct from conventional CD8+ type 1 cytokine producers. The preference for expression of type 1 cytokines displayed by in vivo-activated CD8+ T cells therefore must reflect factors other than precommitment of their naive precursors. We further show that some proliferating CD8+ T cells cultured in type 1-polarizing conditions for many cell divisions retain the potential to respond to IL-4 by expressing type 2 cytokines. This property has implications for strategies that aim to redirect established T cell cytokine responses.
The multipotentiality of naive and activated CD8+ T cells was assessed by comparing the cytokine mRNA profiles of families of short-term subclones activated with anti-receptor antibodies and IL-2 in the presence or absence of exogenous IL-4. There are several features of this protocol that bear on the interpretation of the data.
The first is the use of paired-daughter analysis, rather than the more usual approach of dividing T cell populations into different culture conditions. This allowed us to monitor the fate of every parent cell and every recultured daughter cell, enabling effects on differentiation to be distinguished from effects on survival and proliferation and providing estimates of the frequency of multipotential cells. Several previous studies have examined the stability of murine and human CD4+ T cell IFN-γ/IL-4 profiles under various conditions (4, 28–32) but, because these studies were performed in bulk culture or in vivo, it was not established whether populations or clones whose cytokine profiles changed in different conditions developed by differentiation from persistent multipotential cells or by selective expansion of a subset of committed cells. This difficulty previously has been noted in experiments where dramatic cell death was observed when CD4+ T cells cultured under one set of polarizing conditions were transferred to the opposite conditions (31). In the present study, however, because ≥75% of daughters and granddaughters formed clones whether or not IL-4 was added, and almost 50% of subclones grown in IL-2 + IL-4 expressed IL-4 compared with only 2% of their siblings cultured in IL-2 alone, we can conclude that most daughters and granddaughters were also multipotential at the time of subcloning. Similarly, in analyses of clones expanded for ≥7 divisions, the high recloning efficiencies of individual cells from these clones and their IL-4 responsiveness suggest that the IL-4-responsive cells were multipotential rather than previously committed, selectively expanded cells.
A second feature of the experimental design was the use of anti-receptor antibodies rather than antigen-presenting cells to activate single CD8+ T cells. The absence of any cell other than the T cell enabled us to monitor cell division, identify and transfer individual daughters, and conclude with certainty that T cells were both the target of any stimuli and the source of the detected mRNA. While the strength and duration of stimulation with immobilized antibodies are probably greater than generally achieved with antigen-presenting cells and the combination of receptors ligated may influence the frequency at which each cytokine is expressed, this method of stimulation does not override the potent effect of IL-4 on expression of type 2 cytokine genes. Anti-receptor antibodies thus provided a way to activate most normal CD8+ T cells and reveal their cytokine-producing potential in the presence and absence of IL-4.
Cytokine profiles were determined by RT-PCR 3–5 days after subcloning. This short time-course minimized clonal attrition, maximizing the proportion of parent cells for which paired subclone data were obtained. On the other hand, it probably revealed more diversity among cytokine profiles than would be seen in long-term clones cultured in the same conditions. The data in Fig. 4A suggest such a loss of diversity with longer clonal expansion in IL-2 but the extent to which this reflects preferential survival of cells with a certain phenotype is not known. The small size of the subclones and insensitivity of protein assays for the cytokines studied here, compared with the single-cell sensitivity of RT-PCR, meant that cytokine profiles could not be checked at the protein level. In other systems, however, we have observed strong correlations between the frequencies of cytokine-synthesizing cells or clones detected by RT-PCR and by assays for secreted or intracellular protein in various T cell populations, and a direct correspondence between IL-3 mRNA and protein detection in single cells (refs. 24 and 25; D. R. Fitzpatrick, N. Baumgarth, and A.K., unpublished observations).
Perhaps the most important issue for this study is whether the observed differences between cytokine mRNA combinations detected in sibling clones reflect real differences or simply transient variations in expression patterns due to kinetic or other variables. Two types of evidence support the former conclusion. The first is that the patterns were markedly affected by culture in IL-4, consistent with published results in many different T cell activation systems. Frequencies of IL-4 and IL-6 expression in particular were increased >20-fold by exogenous IL-4. When the analysis of multipotentiality shown in Table 1 is limited to these two cytokines (absence of IL-4 and IL-6 mRNA in the subclone grown in IL-2 and presence of either or both mRNAs in the subclone grown in IL-2 + IL-4), at least 56% of parent cells were multipotential. Second, as reviewed elsewhere (33), several laboratories have shown that individual T cells within polarized clones and populations can display diverse cytokine expression patterns. When the contribution of cell-cell variation is minimized by limiting the analysis to the largest subclone pairs (≥100 cells in each subclone), 96% of parent cells were multipotential (n = 47), compared with 89% for the whole group. Combining these two factors by considering only IL-4 and IL-6 expression by these larger subclones, 81% of parent cells were multipotential. Thus the multipotentiality of most parent CD8+ T cells is apparent even when the impact of single-cell heterogeneity is reduced and the analysis is restricted to the cytokines whose expression is most dependent on exogenous IL-4 and therefore unlikely to be due to transient variations in expression.
Cells able to respond to IL-4 by expressing type 2 cytokines persisted in a substantial proportion of clones for at least 7 or 13–15 cell divisions in IL-2. Because such clones expand exponentially under the conditions used, the probability is high that the IL-4-responsive cells also had undergone many cell divisions. We previously have found that characteristic patterns of granulocyte-macrophage colony stimulating factor and IL-3 secretion also were transmitted to clonal progeny by some, but not all, cells subcloned from primary CD4+ T cell clones of 2–16 cells (21). Multipotentiality was detected at lower frequency in clones assayed after ≥7 divisions than after 1–2 divisions in the present study, but an extensive parallel analysis will be required to establish whether the frequency of multipotential cells does in fact decline with clonal expansion. Others have shown that 7-day CD4+ T cell populations induced to synthesize IFN-γ without IL-4 by culture with IL-12 could be converted to the opposite phenotype by exposure to IL-4 (29–31), but that type 1 CD4+ populations and clones ultimately can lose this ability to alter their cytokine profiles (4, 31). On the other hand and in contrast to the present study, Sad and Mosmann (7, 34) have reported that 6-day populations and 12-day clones of CD8+ T cells with a type 1 profile no longer synthesized IL-2 but otherwise retained their type 1 profile when cultured in IL-4. This difference may partly reflect the use of population splitting, rather than subcloning as in the present study, because the dominant phenotype may prevent expansion or detection of minority types in bulk cultures.
A surprisingly high proportion (36%) of daughters and granddaughters cultured in the absence of exogenous IL-4 gave rise to subclones that expressed one or more type 2 cytokines, particularly IL-10. Production of IL-10 by some CD8+ T cell clones that otherwise displayed a type 1 profile also has been observed by others (7). It is unlikely that the type 2 cytokine producers in the present study were all derived from cells exposed to IL-4 during activation in vivo because parent cells were selected for small size and low expression of the long-lasting activation marker CD44 (26, 27), and most parents of these subclones appeared multipotential. We favor the alternative explanation that de novo expression of IL-4 and other type 2 cytokine genes can occur in the absence of exogenous IL-4, either spontaneously or induced by IL-4-independent pathways. Work is in progress to assess the ability of various costimulatory antibodies to activate IL-4-independent pathways of type 2 cytokine synthesis. The existence of such pathways would bear on the current debate on the origin of IL-4 thought to be required to initiate type 2 cytokine synthesis by T cells in vivo (1, 2), and is suggested by reports that T cells from IL-4−/− and Stat6−/− mice synthesized measurable levels of some type 2 cytokines (35–37), and that naive murine and human CD4+ T cells produced IL-4 when repeatedly stimulated without exogenous IL-4 (38, 39) or immunized after transfer into irradiated IL-4-deficient mice (40). Further work will be needed to determine whether the expression of IL-5 and IL-10 detected in IL-4-negative subclones grown without exogenous IL-4 depended on endogenous, perhaps transient, IL-4 production. Importantly, we have not detected transient IL-4 synthesis either in bulk cultures (20) or during early clonal expansion in the absence of exogenous IL-4; in fact, IL-4 and all the other cytokines measured here were detected more frequently in larger clones and at later time points over the 7-day time frame of these experiments (unpublished observations).
Culture in IL-4 markedly increased the frequencies of IL-4, IL-6 and, to a lesser extent, IL-2, IL-5 and IL-10 expression among the subclones. Unexpectedly in view of earlier studies (6, 8), the frequency of IFN-γ producers was not significantly reduced in the presence of IL-4. This may reflect the short duration of exposure to IL-4 or insufficient cell densities to achieve inhibitory concentrations of crossregulatory cytokines. It is also possible that subclones grown in IL-4 synthesized less IFN-γ than those grown without IL-4 because the RT-PCR method measured whether or not subclones contained detectable cytokine mRNA but did not quantify the amount per subclone.
In conclusion, peripheral CD8+ T cells apparently have as many choices in their expression of cytokine genes as their CD4+ counterparts. While many class I-dependent immune responses favor type 1 cytokine expression in CD8+ cells, the potential diversity of their cytokine profiles and the fact that CD8+ type 2 cytokine producers are sometimes found under physiological conditions suggest that the immunoregulatory roles of CD8+ T cells might also be as diverse as those of CD4+ T cells.
Acknowledgments
We thank Dr. David Fitzpatrick, Prof. Michael Good, Dr. Nick Gough, Prof. Donald Metcalf, Dr. Bill Paul, and Dr. Ranjeny Thomas for invaluable advice and comment, and Diana Battistutta for assistance with statistical analyses. This work was supported by the Australian National Health and Medical Research Council and the Queensland Institute of Medical Research Trust.
ABBREVIATIONS
- IFN-γ
interferon γ
- IL
interleukin
- RT-PCR
reverse transcription–PCR
References
- 1.Seder R A, Paul W E. Annu Rev Immunol. 1994;12:635–673. doi: 10.1146/annurev.iy.12.040194.003223. [DOI] [PubMed] [Google Scholar]
- 2.Abbas A K, Murphy K M, Sher A. Nature (London) 1996;383:787–793. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- 3.Röcken M, Saurat J-H, Hauser C J. J Immunol. 1992;148:1031–1036. [PubMed] [Google Scholar]
- 4.Sad S, Mosmann T R. J Immunol. 1994;153:3514–3522. [PubMed] [Google Scholar]
- 5.Seder R A, Boulay J-L, Finkelman F, Barbier S, Ben-Sasson S Z, Le Gros G, Paul W E. J Immunol. 1992;148:1652–1656. [PubMed] [Google Scholar]
- 6.Erard F, Wild M-T, Garcia-Sanz J A, Le Gros G. Science. 1993;260:1802–1805. doi: 10.1126/science.8511588. [DOI] [PubMed] [Google Scholar]
- 7.Sad S, Marcotte R, Mosmann T R. Immunity. 1995;2:271–279. doi: 10.1016/1074-7613(95)90051-9. [DOI] [PubMed] [Google Scholar]
- 8.Croft M, Carter L, Swain S L, Dutton R W. J Exp Med. 1994;180:1715–1728. doi: 10.1084/jem.180.5.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fong T A T, Mosmann T R. J Immunol. 1990;144:1744–1752. [PubMed] [Google Scholar]
- 10.Kelso A, Troutt A B, Maraskovsky E, Gough N M, Morris L, Pech M H, Thomson J A. Immunol Rev. 1991;123:85–114. doi: 10.1111/j.1600-065x.1991.tb00607.x. [DOI] [PubMed] [Google Scholar]
- 11.Carding S R, Allan W, McMickle A, Doherty P C. J Exp Med. 1993;177:475–482. doi: 10.1084/jem.177.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelso A, Gough N M. Proc Natl Acad Sci USA. 1988;85:9189–9193. doi: 10.1073/pnas.85.23.9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taguchi T, McGhee J R, Coffman R L, Beagley K W, Eldridge J H, Takatsu K, Kiyono H. J Immunol. 1990;145:68–77. [PubMed] [Google Scholar]
- 14.Baumgarth N, Brown L, Jackson D, Kelso A. J Virol. 1994;68:7575–7581. doi: 10.1128/jvi.68.11.7575-7581.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coyle A J, Erard F, Bertrand C, Walti S, Pircher H, Le Gros G. J Exp Med. 1995;181:1229–1233. doi: 10.1084/jem.181.3.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erb K J, Le Gros G. Immunol Cell Biol. 1996;74:206–208. doi: 10.1038/icb.1996.29. [DOI] [PubMed] [Google Scholar]
- 17.Xu H, DiIulio N A, Fairchild R L. J Exp Med. 1996;183:1001–1012. doi: 10.1084/jem.183.3.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metcalf D. Proc Natl Acad Sci USA. 1980;77:5327–5330. doi: 10.1073/pnas.77.9.5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suda T, Suda J, Ogawa M. Proc Natl Acad Sci USA. 1984;81:2520–2524. doi: 10.1073/pnas.81.8.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maraskovsky E, Troutt A B, Kelso A. Int Immunol. 1992;4:475–485. doi: 10.1093/intimm/4.4.475. [DOI] [PubMed] [Google Scholar]
- 21.Fitzpatrick D R, Kelso A. J Immunol. 1995;155:5140–5150. [PubMed] [Google Scholar]
- 22.Groves P L, Pech M H, Troutt A B, Kelso A. Immunology. 1994;83:25–32. [PMC free article] [PubMed] [Google Scholar]
- 23.Smith K G C, Nossal G J V, Tarlinton D M. Proc Natl Acad Sci USA. 1995;92:11628–11632. doi: 10.1073/pnas.92.25.11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Troutt A B, Kelso A. Proc Natl Acad Sci USA. 1992;89:5276–5280. doi: 10.1073/pnas.89.12.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelso A, Groves P, Troutt A B, Francis K. Eur J Immunol. 1995;25:1168–1175. doi: 10.1002/eji.1830250506. [DOI] [PubMed] [Google Scholar]
- 26.Cerottini J-C, MacDonald H R. Annu Rev Immunol. 1989;7:77–89. doi: 10.1146/annurev.iy.07.040189.000453. [DOI] [PubMed] [Google Scholar]
- 27.Sprent J. Cell. 1994;76:315–322. doi: 10.1016/0092-8674(94)90338-7. [DOI] [PubMed] [Google Scholar]
- 28.Swain S L. Immunity. 1994;1:543–552. doi: 10.1016/1074-7613(94)90044-2. [DOI] [PubMed] [Google Scholar]
- 29.Perez V L, Lederer J A, Lichtman A H, Abbas A K. Int Immunol. 1995;7:869–875. doi: 10.1093/intimm/7.5.869. [DOI] [PubMed] [Google Scholar]
- 30.Szabo S J, Jacobson N G, Dighe A S, Gubler U, Murphy K M. Immunity. 1995;2:665–675. doi: 10.1016/1074-7613(95)90011-x. [DOI] [PubMed] [Google Scholar]
- 31.Murphy E, Shibuya K, Hosken N, Openshaw P, Maino V, Davis K, Murphy K, O’Garra A. J Exp Med. 1996;183:901–913. doi: 10.1084/jem.183.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sornasse T, Larenas P V, Davis K A, de Vries J E, Yssel H. J Exp Med. 1996;184:473–483. doi: 10.1084/jem.184.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelso A. Immunol Today. 1995;16:374–379. doi: 10.1016/0167-5699(95)80004-2. [DOI] [PubMed] [Google Scholar]
- 34.Sad S, Mosmann T R. J Exp Med. 1995;182:1505–1515. doi: 10.1084/jem.182.5.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kopf M, Le Gros G, Bachmann M, Lamers M C, Bluethmann H, Köhler G. Nature (London) 1993;362:245–248. doi: 10.1038/362245a0. [DOI] [PubMed] [Google Scholar]
- 36.Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, Kashiwamura S, Nakanishi K, Yoshida N, Kishimoto T, Akira S. Nature (London) 1996;380:627–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 37.Shimoda K, van Deursen J, Sangster M Y, Sarawar S R, Carson R T, Tripp R A, Chu C, Quelle F W, Nosaka T, Vignali D A A, Doherty P C, Grosveld G, Paul W E, Ihle J N. Nature (London) 1996;380:630–633. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- 38.Croft M, Swain S L. J Immunol. 1995;154:4269–4282. [PubMed] [Google Scholar]
- 39.Demeure C E, Yang L-P, Byun D G, Ishihara H, Vezzio N, Delespesse G. Eur J Immunol. 1995;25:2722–2725. doi: 10.1002/eji.1830250950. [DOI] [PubMed] [Google Scholar]
- 40.Schmitz J, Thiel A, Kühn R, Rajewsky K, Müller W, Assenmacher M, Radbruch A. J Exp Med. 1994;179:1349–1353. doi: 10.1084/jem.179.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]