Abstract
V(D)J recombination requires both lymphoid-specific and generally expressed enzymatic activities. All three known generally expressed activities involved in V(D)J recombination are also involved in DNA double-strand break repair (DSBR). Two of these are components of the DNA-dependent protein kinase (DNA-PK) and include Ku80 and DNA-PK catalytic subunit (DNA-PKcs); the third, XRCC4, is a protein of unknown function. The Ku70 protein is an additional component of DNA-PK; Ku70 forms a heterodimer with Ku80 to generate the DNA end-binding component of the enzyme. To test putative functions for Ku70, we have used gene-targeted mutation to generate a murine embryonic stem cell line which lacks Ku70 expression. We find that the Ku70−/− cells produce no detectable Ku70 and very little Ku80, suggesting a direct interrelationship between their levels. Correspondingly, these cells lack the nonspecific DNA end-binding activity associated with Ku. Significantly, the Ku70−/− embryonic stem cells have markedly increased sensitivity to γ-irradiation relative to Ku70+/− or wild-type embryonic stem cells. Furthermore, the Ku70−/− cells lack the ability to effectively rejoin signal and coding ends liberated in transiently introduced V(D)J recombination substrates by enforced RAG-1 and RAG-2 expression. We conclude that the Ku70 gene product is involved in DSBR and V(D)J recombination and confirm that the Ku70 gene can be classified as a member of the x-ray cross-complementation group 6 (XRCC6). Potential differences between the Ku70−/− and Ku80−/− V(D)J recombination defects are discussed.
Immunoglobulin (Ig) and T cell receptor (TCR) variable region diversity is generated by the V(D)J recombination process, whereby the variable region genes are assembled from multiple germ-line V, (D), and J segments during early B and T cell differentiation (1, 2). Each germ-line V, (D), and J segment is flanked by recombination signal sequences (RSs) that target the V(D)J recombinase activity. Initiation of the reaction involves the introduction of double-strand DNA breaks (DSBs) between the potential V, (D), or J coding sequence and their flanking RSs, resulting in the liberation of RSs as blunt 5′-phosphorylated DSBs and the coding ends in the form of hairpin structures (2, 3). This initial phase of the reaction is carried out by the RAG-1 and RAG-2 proteins (4, 5), which appear to be the only required tissue-specific components of the V(D)J recombinase and which are directly involved in RS recognition and induction of initial cleavage at RS/coding boundaries (6–9). Completion of the V(D)J recombination reaction requires the participation of a number of generally expressed cellular factors. To date, all such factors identified are involved in general DSB repair (DSBR) (10, 11). In this regard, the completion of the V(D)J recombination reaction can also be viewed as a DSBR process in which generally expressed factors are recruited to repair the specifically broken ends generated by RAG-1 and RAG-2 activity.
The three known gene products that are required for the DSBR portion of the V(D)J recombination reaction were identified through studies of ionizing-radiation-sensitive Chinese hamster cell lines and cells from severe combined immune-deficient (Scid) mice (10). There are at least eight x-ray cross-complementation (XRCC) groups, based on somatic cell hybrid studies (12, 13). Of these, groups XRCC4, XRCC5, and XRCC7, which have specific defects in DSBR (14–17), were also found to have defects in their ability to support V(D)J recombination (18–20). In this regard, following introduction of RAG-1 and RAG-2 gene expression, cell lines from each complementation group were capable of initiating V(D)J recombination on transiently introduced V(D)J recombination substrates. However, XR-1 and xrs-6 cells (representing the XRCC4 and -5 groups, respectively) were defective in their ability to form both coding and RS joins (18, 19), whereas V3 cells (from the XRCC7 group) had impaired ability to form coding joins but were relatively normal in ability to form RS joins (18, 21). V3 cells were further shown to represent the same genetic complementation group as the murine Scid mutation (20).
xrs-6 cells also were found to lack a DNA end-binding activity associated with the Ku complex (22–24). The Ku complex was first identified using autoantibodies isolated from patients with systemic lupus erythematosus and consists of a DNA end-binding heterodimer of 70-kDa (Ku70) and 80-kDa (Ku80) subunits (25, 26). Expression of a transfected human Ku80 gene restored radiosensitivity, DNA end-binding activity, and ability to support V(D)J recombination to xrs-6 cells (22, 27, 28). In addition, debilitating mutations in the Ku80 gene were identified both in xrs-6 cells and in other complementation group 5 cells (29–31). Thus, Ku80 is the XRCC5 gene product. As predicted, B and T cell development in Ku80-deficient mice is arrested at an early stage, accompanied by defects in both RS and coding join formation during attempted V(D)J recombination (32, 33). Additionally, the Ku80-deficient mice are smaller than their wild-type or heterozygous siblings, and Ku80-deficient cells show premature senescence, suggesting a link between Ku80 and growth control (32).
The Ku heterodimer was also shown to be the DNA-binding component of an enzyme referred to as the DNA-dependent protein kinase (DNA-PK), the catalytic subunit of which is approximately 450 kDa and referred to as DNA-PKcs (34). xrs-6 cells are defective in DNA-PK activity (35) due to the lack of Ku80 expression (22, 27). V-3 cells express normal levels of the Ku complex but were found to be deficient in expression of the DNA-PKcs component (17, 36). Expression of the DNA-PKcs gene corrected the DSBR and V(D)J recombination defects in V-3 cells (17, 36), and the DNA-PKcs and scid genes were mapped to the same chromosomal regions (17, 37–39), strongly indicating that the DNA-PKcs is the XRCC7 gene product. In support of this conclusion, a nonsense mutation was identified in the carboxyl-terminal region of DNA-PKcs in murine Scid cells (40, 41).
As two of the three known components of DNA-PK have been implicated in mutations that affect DSBR and V(D)J recombination, the Ku70 gene also was proposed to be required for these processes and tentatively designated the XRCC6 gene (42). However, to date, no Ku70 mutation has been isolated from any of the XRCC groups. Indeed, the XRCC4 gene (deficient in XR-1 cells) has been identified through a complementation cloning approach and found to encode a ubiquitously expressed protein with no homology to any known protein (43). Furthermore, an independent group of ionizing radiation-sensitive Chinese hamster ovary (CHO) cell mutants (sxi-1, -2, and -3) that had combined defects in DSBR and ability to support V(D)J recombination were shown to have defects associated with Ku80 gene expression (28, 44). Thus, the requirement for Ku70 in V(D)J recombination and DSBR has remained speculative. To unequivocally elucidate the role of Ku70 in these processes, we have generated and analyzed Ku70-deficient embryonic stem (ES) cells.
MATERIALS AND METHODS
Ku70 Gene Targeting and Generation of Ku70-Deficient ES Cell Lines.
A full-length cDNA clone for mouse Ku70 gene was isolated from a pre-B cell (18.8) expression library (H. Cheng and F.W.A., unpublished data). The full-length Ku70 cDNA fragment was used as a probe to screen a 129/sv mouse genomic library (Stratagene). To make the Ku70 targeting construct a 5.6-kb SacI–HindIII genomic fragment containing exons 5–7 was modified to a SalI/SacI–HindIII/ClaI/SalI fragment by transferring through two different cloning vectors, and then inserted into the SalI site of pSalI-loxP-neor-loxP-XhoI-tk cassette vector, or pLNtk (45). An exon 3-containing 3.6-kb SalI–SalI genomic fragment from one end of a phage clone was inserted into the XhoI site of the pLNtk vector.
ES cells (J1 line, provided by R. Jaenisch, Massachusetts Institute of Technology) were transfected with 25 μg of ClaI-linearized Ku70 targeting vector DNA and then selected in ES cell medium containing G418 and ganciclovir (generously provided by Syntex) as described previously (46, 47). Homologous recombinants (Ku70+/− cells) were identified by Southern blot analysis of BamHI-digested DNA probed with an 850-bp SacI fragment from mouse Ku70 cDNA. To select for Ku70−/− cells, we applied high concentrations of G418, as previously described (48), on Ku70+/− cells. Using the same Southern blot analysis, we identified four clones (two from each Ku70+/− clone) as Ku70−/− clones as judged by the disappearance of the wild-type restriction fragment. To remove the murine embryonic fibroblasts (MEFs), ES cells, including wild-type (J1), Ku70+/−, and Ku70−/− cells, were trypsinized, seeded on gelatinized plates, and incubated for 30 min. After repeating the same procedure for depleting MEFs, the ES cells were seeded on a new gelatinized plate for growth. These MEF-free ES cells were then used for characterization.
Western Blot Analysis.
Two million ES cells were lysed in SDS/sample buffer and boiled for 5–10 min at 100°C. After a brief spinning, supernatants were directly resolved by SDS/PAGE, then transferred to a poly(vinylidene difluoride) (PVDF) membrane (NEN), and probed with either goat anti-mouse Ku70 or goat anti-mouse Ku80 antiserum (Santa Cruz Biotechnology) followed by horseradish peroxidase-conjugated protein A+G. Enhanced chemiluminescence detection (ECL, Amersham) was used to develop the results.
Determination of Radiosensitivity by a Colony Survival Assay.
MEF-free ES cells were trypsinized and resuspended in ES cell medium. After exposure to different doses of a 137Cr source in a γ-irradiator, the ES cells were seeded onto gelatinized plates. After 7 days, the colonies were stained on the plates with crystal violet (Kodak) and counted. The numbers of surviving colonies at different dosages were calculated as the average of triplicates.
DNA End-Binding Assay.
A 32P-labeled NotI–XhoI fragment of pBluescript SK II (+) (Stratagene) was prepared. Closed circular pBluescript SK II (+) DNA was used as a DNA competitor. Preparation of nuclear extracts from ES cells, DNA end-binding assays, and mobility shift gels were prepared by following previously described procedures (23, 28).
Transient V(D)J Recombination Assay.
Recombination substrates pJH200 and pJH290 were used in the transient V(D)J recombination assay to test for RS and coding joins, respectively, as previously described in various cell types (49, 20). For assay in ES cells, minor modifications were applied to enhance the transfection efficiency. Briefly, MEF-free ES cells (about 5 × 106) were seeded onto gelatinized 10-cm plates 7 hr before the transfection. One to 2 μg of the pJH200 or pJH290 recombination substrates was cotransfected with 3–4 μg of genomic RAG-1 and RAG-2 sequences under the control of the Rous sarcoma virus long terminal repeat promoter by a standard calcium phosphate method. Extrachromosomal DNA was recovered after 48 hr by an alkaline lysis method and electroporated into Escherichia coli MC1061. V(D)J recombination activity was measured by comparing the ratio of ampicillin-resistant (Ampr) chloramphenicol-resistant (Camr) colonies versus Ampr colonies among ES cells with different genotypes. Because a precise signal join generates an ApaLI site, ApaLI digestion of PCR products from the recombinant substrate pJH200 was used to measure the fidelity of the signal joins. The coding joins of recovered recombinant substrate pJH290 were also analyzed by nucleotide sequencing.
RESULTS
Generation of Ku70-Deficient ES Cell Lines.
To understand the role of Ku70 in V(D)J recombination and in DSBR, we generated Ku70-deficient ES cells. A full-length cDNA clone encoding the mouse Ku70 gene was isolated and used to screen a mouse genomic library. Five genomic clones isolated from this library were characterized. Localization of exons was determined by hybridization of Ku70 cDNA fragments to the genomic clones (Fig. 1A) and generated a partial map of the murine Ku70 genomic locus that corresponded to that determined by a recently published study (50). Coding exons 2–10 and their exon/intron boundaries were confirmed by nucleotide sequence analysis (data not shown). It was previously suggested that a family of genes encoding Ku70-related proteins might exist in humans, on the basis of Southern blot analysis under different hybridization stringencies (51). However, our mapping and nucleotide sequence data indicated that the five Ku70 hybridizing clones were overlapping and represented a single genomic locus (data not shown).
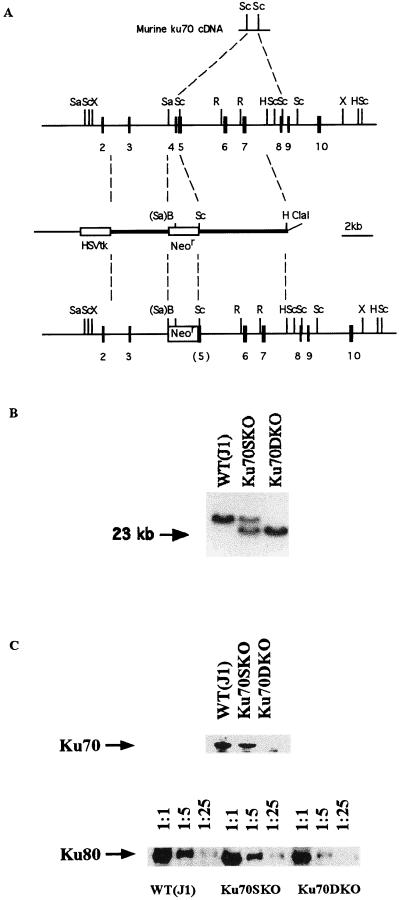
Figure 1.
Ku70 gene targeting. (A) Schematic diagrams of the murine Ku70 cDNA, partial genomic map for the Ku70 gene, Ku70 targeting construct, and Ku70 targeted genomic locus. B, BamHI; H, HindIII; R, EcoRI; Sa, SalI; Sc, SacI; X, XhoI; and Neor, neomycin-resistance gene. (B) Typical result of the Southern blot analysis of BamHI-digested DNA prepared from Ku70 mutant ES cells. The 32P-labeled probe used was a SacI fragment from 3′ of murine Ku70 cDNA that is coded by exons located outside the 3′ genomic fragment for the construct. The lower band corresponds to the targeted allele and the upper band represents the untargeted allele. The Ku70−/− (Ku70DKO) ES cells were identified by the Southern blot analysis as judged by disappearance of the upper band. (C) Western blot analysis of Ku70-deficient ES cells. Total cell lysates were prepared from the MEF-free ES cells. To detect the protein level of Ku80, cell lysates were diluted 1:1, 1:5, and 1:25, and resolved by SDS/PAGE before Western blotting. Anti-mouse Ku70 antibody and anti-mouse Ku80 antibody were used for detection.
The various activities associated with the Ku70/Ku80 protein complex have not been dissected into structural domains. Therefore, to inactivate Ku70 function, we created a targeting vector aiming to delete a 450-bp SalI–SacI fragment containing exon 4 and part of exon 5 and to replace this region with a neomycin-resistance (Neor) gene (Fig. 1A). A potential transcript generated from the targeting event would have a reading frame shift that would lead to a truncated protein of only the first 64 of the total 608 amino acids. J1 ES cells were transfected with linearized vector and selected with G418 and ganciclovir as described previously (46, 47). Homologous recombinants were identified by Southern blotting of BamHI-digested DNA probed with an 850-bp SacI fragment from mouse Ku70 cDNA. Thirteen of 109 ES cell clones displayed a targeted band of 24 kb and a wild-type band of over 40 kb (a typical result is indicated in Fig. 1B). To further analyze whether any aberrant recombination events occurred in these clones, DNAs were digested with SacI and HindIII, respectively, and probed either with a 400-bp SacI fragment from the 5′ region of Ku70 cDNA or with a probe from a Neor gene fragment (data not shown). The results of these analyses confirmed that all 13 Ku70+/− ES cell clones harbored the desired targeted mutagenesis event (shown in Fig. 1A). To generate Ku70−/− ES cells, 2 Ku70+/− clones, Ku70SKO29 and Ku70SKO34, were further cultured in increased concentrations G418 as described previously (48). Ku70−/− ES cells were identified by Southern blotting analyses using the same probe as for identifying Ku70+/− clones. Four independent Ku70−/− clones were obtained at 4 mg/ml G418 (a typical result is shown in Fig. 1B).
Ku70−/− ES cells were indistinguishable from wild-type ES cells and Ku70+/− ES cells when grown on MEF feeder cells. Like wild-type J1 and Ku70+/− ES cells, Ku70−/− ES cells could also be grown on gelatinized plates without MEF feeder cells with no obvious morphological differences. To further confirm that Ku70−/− ES cells indeed lacked expression of the Ku70 protein, we performed a Western blot analysis of wild-type, Ku70+/−, and Ku70−/− ES cells. While Ku70 protein was clearly detectable in wild-type ES cells and Ku70+/− ES cells, we did not detect any Ku70 protein in Ku70−/− ES cells when we used an anti-mouse Ku70 antibody (Fig. 1C). Thus, expression of the Ku70 protein is not essential for general cell maintenance, survival, and propagation in the absence of DNA-damaging agents. Previous studies have also indicated that Ku70 expression is reduced in Ku80-deficient cells (30, 31). To determine if the reverse also holds, we used Western blotting procedures to assay for Ku80 expression in the cell lines described above and found that its protein level was reduced to approximately  compared with that of control ES cells (Fig. 1C)
compared with that of control ES cells (Fig. 1C)
Ku70-Deficient ES Cells Lose Nonspecific Double-Strand DNA End-Binding Activity.
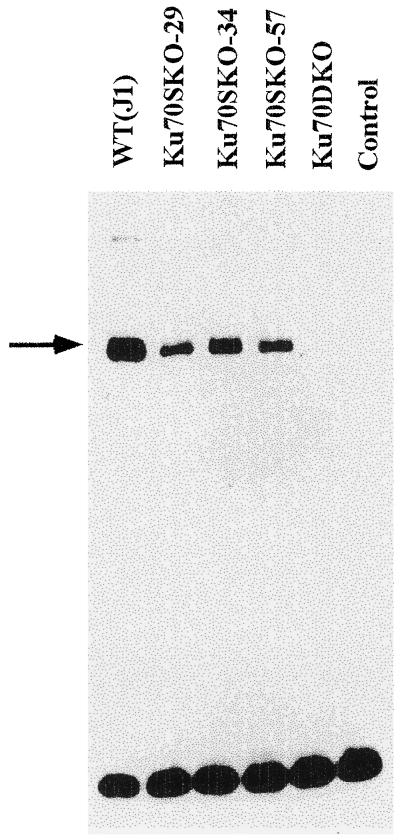
The Ku70/80 complex binds to DNA double-strand ends in vitro, and loss of this activity is observed in cell extracts from Ku80-deficient cells. To test whether loss of Ku70 expression similarly depleted DNA double-strand end-binding activity, we prepared whole cell nuclear extracts from ES cells that had been grown several passages without MEFs. The extracts were then incubated with a 32P-labeled 70-bp DNA fragment, and binding was measured by a electrophoretic mobility-shift assay (23, 24). Wild-type ES cells (Fig. 2, lane 1) displayed a gel shift pattern similar to the patterns from other cell types, which was defined as a major B1 band and a faint B2 band with slower mobility (23). In contrast, nuclear extracts prepared from the Ku70−/− cells generated no shifted band (Fig. 2, lane 5), even after prolonged exposure of the gel (data not shown). Of note, extracts from three independent Ku70+/− cells displayed a distinct gel-shift pattern (Fig. 2, lanes 3, 4, and 5) with reduced levels of the shifted complex relative to wild-type J1 cell extracts, possibly reflecting a gene dosage effect on Ku70 protein levels.
Figure 2.
Ku70-deficient ES cells lose DNA end-binding activities as measured by gel mobility-shift assay. Wild-type (J1), three Ku70+/− (Ku70SKO-29, Ku70SKO-34, and Ku70SKO-57), and Ku70−/− (Ku70DKO) ES cells were tested for their DNA end-binding activities. Nuclear extracts prepared from different ES cells as indicated were incubated with a 32P-labeled probe and resolved on the gel. The control lane contains the 32P-labeled probe without incubation with any nuclear extract. Wild-type (J1) ES cells display a gel-shift pattern similar to other cell types described (29). The arrow points to the shifted band, or B1 band. Ku70−/− (Ku70DKO) ES cells show no shifted band even after prolonged exposure time.
Ku70-Deficient ES Cells Are Sensitive to γ-Irradiation.
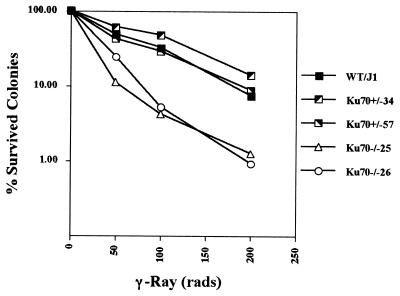
The XRCC group cells were established by screening for a radiation-sensitive phenotype. It seemed likely, based on its known functions, that Ku70 deficiency may also lead to increased sensitivity to ionizing radiation similar to Ku80 or DNA-PKcs deficiency. Therefore, we tested the radiosensitivity of wild-type, Ku70+/−, and Ku70−/− ES cells by using a colony survival assay. The ES cells passaged without MEFs were exposed to various doses of γ-irradiation before being seeded onto gelatinized plates. Surviving colonies were counted 7 days after irradiation. Analysis of three independent Ku70+/− clones and two independent Ku70−/− clones indicated that the Ku70−/− cells were markedly sensitive to γ-irradiation as compared with Ku70+/− cells, whose radiation sensitivity was indistinguishable from that of the parental ES cells (Fig. 3).
Figure 3.
Ku70-deficient cells have increased sensitivity to γ-irradiation. Wild-type (J1), two Ku70+/− (KuSKO34 and KuSKO57), and two Ku70−/− (KuDKO25 and KuDKO26) ES cells were exposed to γ-irradiation at doses of 50, 100, or 200 rads and were seeded on gelatinized plates. Surviving colonies were counted on the seventh day after irradiation. Each point in this plot is the average of triplicates and is a percentage of the colony counts from unirradiated ES cells.
Ku70 Deficiency Affects Both Signal and Coding Joining in a Transient V(D)J Recombination Assay.
Previous analyses have shown that Ku80-deficient cells have defects in ability to support the V(D)J recombination reaction. To test the possibility that Ku70 may also participate in this reaction, we assayed V(D)J recombination by providing Ku70−/− and control ES cells with RAG-1 and RAG-2 by means of transient transfection of appropriate expression vectors, and we assayed the ability of these RAG-transfected cells to rearrange transient V(D)J recombination constructs (19). The transient recombination substrates used, pJH200 and pJH290 (49), allowed us to assess RS and coding join formation, respectively. The transient tranfection assay was performed in wild-type, Ku70+/−, and Ku70−/− ES cells. At least four independent transfection experiments were carried out with each line (Table 1). For both RS and coding join substrates, the percentage of recombined products recovered from wild-type ES cells indicated by (Ampr + Camr)/Ampr was significant (Table 1) and comparable to that obtained from other cell lines previously employed in these V(D)J recombination assays (e.g., see refs. 19 and 49). Thus, like other tested cells, ES cells possess all of the generally expressed activities required for V(D)J recombination.
Table 1.
Ku70-deficient ES cells fail to support both signal and coding joining in V(D)J recombination
| Cell line | pJH200 (signal joining)
|
pJH290 (coding joining)
|
|||||
|---|---|---|---|---|---|---|---|
| (Ampr + Camr)/Ampr | % | Relative level | Fidelity, % | (Ampr + Camr)/Ampr | % | Relative level | |
| Exp. I | |||||||
| Wild type (J1) | 144/6,970 | 2.1 | 1.0 | 100 (4/4) | 51/5,270 | 0.97 | 1.0 |
| Ku70SKO | 267/32,300 | 0.83 | 0.4 | 100 (4/4) | 93/9,520 | 0.98 | 1.0 |
| Ku70DKO | 1/17,510 | 0.0057 | 0.0027 | 0 (0/1) | 7/6,800 | 0.1 | 0.1 |
| Exp. II | |||||||
| Wild type (J1) | 24/9,690 | 0.25 | 1.0 | 100 (6/6) | 1,132/251,600 | 0.45 | 1.0 |
| Ku70SKO | 49/29,920 | 0.16 | 0.64 | 100 (6/6) | 104/42,330 | 0.25 | 0.55 |
| Ku70DKO | 0/13,430 | <0.007 | <0.02 | — | 5/41,140 | 0.012 | 0.027 |
| Exp. III | |||||||
| Wild type (J1) | 136/96,000 | 0.14 | 1.0 | 100 (6/6) | 99/29,600 | 0.45 | 1.0 |
| Ku70SKO | 66/75,200 | 0.088 | 0.63 | 100 (6/6) | 47/21,600 | 0.22 | 0.67 |
| Ku70DKO | 0/5,950 | <0.01 | <0.07 | — | 7/206,400 | 0.0034 | 0.01 |
| Exp. IV | |||||||
| Wild type (J1) | 105/6,720 | 1.56 | 1.0 | 100 (6/6) | 262/37,500 | 0.7 | 1.0 |
| Ku70SKO | 148/17,880 | 0.83 | 0.53 | 100 (6/6) | 279/44,040 | 0.63 | 0.9 |
| Ku70DKO | 2/24,210 | 0.0083 | 0.0053 | 0 (0/2) | 3/13,230 | 0.023 | 0.03 |
In contrast to wild-type ES cells, Ku70−/− ES cells have severe defects in supporting V(D)J recombination. First, the frequency of recovered RS joins in Ku70−/− ES cells was reduced to approximately 1% or less relative to that in wild-type ES cells (Table 1). In fact, RS joins were recovered in only two of four experiments; and none of those recovered were precise, as indicated by resistance to digestion with the ApaLI enzyme. In Ku70+/− ES cells, levels of RS appeared to be slightly, but consistently, reduced compared with those of wild-type ES cells (Table 1). The apparent reduction in RS joining in Ku70+/− ES cells may also be caused by a gene dosage effect on Ku70 protein levels, consistent with the findings of the DNA end-binding assays (Fig. 2).
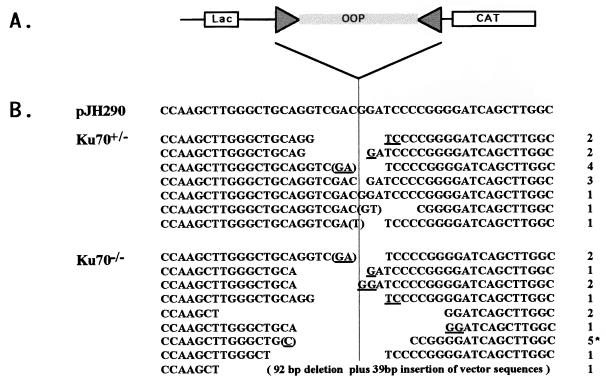
While Ku70+/− ES cells were able to undergo coding join formation at levels comparable to those of wild-type ES cells, the level of coding join formation in Ku70−/− ES cells was markedly decreased from 10- to 100-fold (Table 1). We analyzed the nucleotide sequence of coding joins recovered from Ku70−/− ES cells and compared them to those of coding joins from Ku70+/− ES cells. Ku70+/− coding joins showed characteristics of normal coding joins, including small nucleotide deletions (less than 10 bp) from one or both coding ends, P elements, and occasional redundant nucleotides that may have resulted form homology-mediated joining (Fig. 4B). On average, coding joins rescued from Ku70−/− ES cells had normal characteristics, although they tended to have somewhat larger deletions than those from Ku70+/− ES cells (Table 1). Thus, coding join formation is also severely impaired in Ku70−/− ES cells, but these cells do have ability to make rare junctions that resemble “normal” coding joins.
Figure 4.
Sequence analysis of coding joins rescued from Ku70−/− ES cells in V(D)J recombination assay. (A) Diagram of a region in the pJH290 substrate vector that contains coding sequences. “Lac” box represents the lac operon promoter. “CAT” box represents the chloramphenicol acetyltransferase gene. The CAT gene is expressed only after the transcription terminator (OOP) is removed by recombination involving the joining of RSs (triangles). (B) Sequence of coding joins. In pJH290 coding sequence is assembled by “cut and join.” In Ku70+/−, coding sequences are obtained from randomly picked Ampr Camr clones recovered from Ku70+/− ES cells. In Ku70−/−, coding sequences come from 16 Ampr Camr clones recovered from Ku70−/− ES cells. The number on the right column indicates the number of duplicate sequences recovered from the transfection experiments; all came from different experiments except the five marked by an asterisk, which came from the experiment I in Table 1. Ambiguous nucleotides are underlined. Nucleotides with parentheses can be P elements.
DISCUSSION
To assess the role of Ku70 in DSBR and V(D)J recombination, we have generated a Ku70−/− ES cell line. We show that the Ku70−/− ES cells produce no detectable Ku70 protein, lack the DNA end-binding activities associated with the Ku heterodimer, display increased sensitivity to γ-irradiation relative to wild-type ES cells, and are greatly impaired in ability to form either RS or coding joins in a V(D)J recombination substrate assay. Thus, the observed phenotypes of the Ku70−/− ES cells clearly resemble those of cells lacking Ku80 expression (19, 22, 27). These findings indicate that Ku70, like Ku80, is involved both in general DSBR processes and in the repair of DSBs generated during the V(D)J recombination reaction. Transfection of human Ku70 cDNA expression constructs rescued these defects in Ku70−/− cells (Y. Gu and F.W.A., unpublished observations; S.J. and D.T.W., unpublished observations), allowing us to demonstrate that the Ku70 gene represents the XRCC6 gene as previously proposed (42).
Potential functions for the Ku complex and the DNA-PKcs in the DSBR and V(D)J reactions have been widely discussed (11, 17, 22–24, 27, 28, 32–35, 40, 41, 52). Activation of DNA-PK is likely one of the initial steps of the cellular response to DSBs in their chromosomal DNA, and subsequent phosphorylation of DNA-PK substrates may trigger the repair mechanism (52). The Ku70/80 heterodimer, in addition to its role as the DNA-binding component of DNA-PK, has a number of additional activities, including its nonspecific DNA end-binding activity (23, 24), ATPase and DNA helicase activities (53, 54), and sequence-specific DNA-binding activities (55). In this regard, Ku80-deficient and Ku70-deficient cells are strikingly impaired in ability to form RS joins as compared with DNA-PKcs-defective (Scid) cells (19–21, 33). In addition, Ku80-deficient mice display impaired growth and premature senescence phenotypes not observed in Scid mice. It is currently not known whether these differences are due to additional functions of the Ku80 and Ku70 proteins beyond their role in the DNA-PK complex (17) or due to the characterized DNA-PKcs deficiencies of Scid and V-3 not being complete (41).
The Ku70-deficient and Ku80-deficient phenotypes are similar with respect to their effects on ionizing radiation sensitivity and V(D)J recombination in cell lines. One explanation for the similarities is that the Ku70 and Ku80 proteins perform most, if not all, of their DSBR and V(D)J recombination functions as Ku heterodimers. However, we must note that Ku80 protein levels are substantially decreased in Ku70-deficient ES cells (Fig. 1C). Likewise, previous studies have shown that the Ku70 protein is barely detectable in most Ku80-deficient cells, even though the Ku70 mRNA level was found to be normal (44). Furthermore, normal levels of Ku70 protein can be restored by transfection of Ku80 expression constructs into such cells, suggesting that the Ku80 protein stabilizes the Ku70 protein level (30, 31, 44). Therefore, the Ku70 and Ku80 proteins appear to play reciprocal roles in stabilizing each other, possibly through heterodimer formation. Thus, it remains possible that some aspects of the overlapping phenotype of Ku70 or Ku80 deficiency on V(D)J recombination or DSBR may be due to the loss of an activity associated with just one or the other of the subunits.
There are several potential differences with respect to ability to rearrange transient V(D)J recombination substrates when Ku70-deficient murine ES cells are compared with previously assayed Ku80-deficient CHO cells. With respect to RS join formation, Ku70−/− ES cells appear to be even more severely impaired than Ku80-deficient CHO lines (19, 20, 22, 27). In addition, we were able to rescue coding joins in Ku70−/− ES cells, whereas this was not achieved in Ku80-deficient CHO cells (22). Also, many of the rare coding joins recovered from Ku70−/− ES cells appeared to fall within the normal limits (Fig. 4). In this regard, it is notable that rare coding joins can be recovered from Scid cells, and some of these also appear relatively normal (56–59). The occurrence of such joins contributes to the generation of some mature lymphocytes, resulting in the “leakiness” of the Scid phenotype (60). Consistent with the apparent ability of Ku70−/− ES cells to form some V(D)J coding joins in transient recombination substrates, Ku70-deficient mice, unlike Ku80-deficient mice (32, 33), appear capable of generating some normal mature lymphocytes in vivo (Y. Gu and F.W.A., unpublished results). Therefore, it appears possible that lack of Ku70 versus Ku80 could have subtle differences with respect to end-joining, although this possibility will need to be confirmed through comparison of the Ku80 and Ku70 deficiency in the same cell types.
Acknowledgments
We thank Hwei-Ling Cheng for advice and reagents. We thank Dr. Albert Shaw for reading the manuscript. We also thank Drs. Guillermo Taccioli, Penny Jeggo, and Stephen Jackson for critically evaluating this manuscript. This work was supported by the Howard Hughes Medical Institute and by National Institutes of Health Grants AI20047, AI315714, and CA42335 to F.W.A and by National Institutes of Health Grant CA54326 and a Sandoz Drug Discovery Grant to D.T.W. Y. Gu is a Fellow of the Cancer Research Institute. S.J. is supported by a National Research Service Award Postdoctoral Fellowship. Y. Gao is supported by a postdoctoral fellowship from the Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation.
ABBREVIATIONS
- ES cells
embryonic stem cells
- DSB
double-strand DNA break
- DSBR
DSB repair
- XRCC
x-ray cross-complementation
- RS
recombination signal
- Scid
severe combined immune-deficient
- DNA-PK
DNA-dependent protein kinase
- DNA-PKcs
DNA-dependent protein kinase catalytic subunit
- MEF
murine embryonic fibroblast
- CHO
Chinese hamster ovary
- Ampr
ampicillin-resistant
- Camr
chloramphenicol-resistant
References
- 1.Tonegawa S. Nature (London) 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 2.Okada A, Alt F W. In: Immunoglobulin Genes. 2nd Ed. Honjo T, Alt F W, editors. London: Academic; 1995. pp. 205–234. [Google Scholar]
- 3.van Gent D C, McBlane J F, Ramsden D A, Sadofsky M J, Hesse J E, Gellert M. In: Molecular Analysis of DNA Rearrangements in the Immune System. Jessberger R, Lieber M R, editors. Berlin: Springer; 1996. pp. 1–10. [Google Scholar]
- 4.Schatz D G, Oettinger M A, Baltimore D. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 5.Oettinger M A, Schatz D G, Gorka C, Baltimore D. Science. 1990;24:1517–1522. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 6.van Gent D C, McBlane J F, Ramsden D A, Sadofsky M J, Hesse J E, Gellert M. Cell. 1995;81:925–934. doi: 10.1016/0092-8674(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 7.McBlane J F, van Gent D C, Ramsden D A, Romeo C, Cuomo C A, Gellert M, Oettinger M A. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 8.Difilippantonio M J, McMahan C J, Eastman Q M, Spanopoulou E, Schatz D G. Cell. 1996;87:253–262. doi: 10.1016/s0092-8674(00)81343-4. [DOI] [PubMed] [Google Scholar]
- 9.Spanopoulou E, Zaiteva F, Wang F-H, Santagata S, Baltimore D, Panayotou G. Cell. 1996;87:263–276. doi: 10.1016/s0092-8674(00)81344-6. [DOI] [PubMed] [Google Scholar]
- 10.Taccioli G E, Alt F W. Curr Opin Immunol. 1995;7:436–440. doi: 10.1016/0952-7915(95)80085-9. [DOI] [PubMed] [Google Scholar]
- 11.Weaver D T. Trends Genet. 1995;11:388–392. doi: 10.1016/s0168-9525(00)89121-0. [DOI] [PubMed] [Google Scholar]
- 12.Jones N J, Cox R, Thacker J. Mut Res. 1988;193:139–144. doi: 10.1016/0167-8817(88)90044-2. [DOI] [PubMed] [Google Scholar]
- 13.Jeggo P A, Tesmer J, Chen D J. Mut Res. 1991;254:125–133. doi: 10.1016/0921-8777(91)90003-8. [DOI] [PubMed] [Google Scholar]
- 14.Giaccia A J, Weistein R, Hu J, Stamato T D. Somat Cell Mol Genet. 1985;11:485–491. doi: 10.1007/BF01534842. [DOI] [PubMed] [Google Scholar]
- 15.Jeggo P A, Kemp L M. Mut Res. 1983;112:313–327. doi: 10.1016/0167-8817(83)90026-3. [DOI] [PubMed] [Google Scholar]
- 16.Whitmore G F, Varghese A J, Gulyas S. Int J Radiat Biol. 1989;56:657–665. doi: 10.1080/09553008914551881. [DOI] [PubMed] [Google Scholar]
- 17.Blunt T, Finnie N J, Taccioli G E, Smith G C M, Demengeot J, Gottlieb T M, Mizuta R, Varghese A J, Alt F W, Jeggo P A, Jackson S P. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 18.Taccioli G E, Rathbun G, Shinkai Y, Oltz E M, Cheng H, Whitmore G, Stamato T, Jeggo P, Alt F W. Curr Top Microbiol Immunol. 1992;182:107–114. doi: 10.1007/978-3-642-77633-5_13. [DOI] [PubMed] [Google Scholar]
- 19.Taccioli G E, Rathbun G, Oltz E, Stamato T, Jeggo P A, Alt F W. Science. 1993;260:207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- 20.Taccioli G E, Cheng H-L, Varghese A J, Whitmore G, Alt F W. J Biol Chem. 1994;269:1–4. [PubMed] [Google Scholar]
- 21.Pergola F, Zdzienicka M Z, Lieber M R. Mol Cell Biol. 1993;13:3464–3471. doi: 10.1128/mcb.13.6.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taccioli G E, Gottlieb T M, Blunt T, Priestley A, Demengeot J, Mizuta R, Lehmann A R, Alt F W, Jackson S P, Jeggo P A. Science. 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 23.Rathmell W K, Chu G. Mol Cell Biol. 1994;14:4741–4748. doi: 10.1128/mcb.14.7.4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Getts R C, Stamato T D. J Biol Chem. 1994;269:15981–15984. [PubMed] [Google Scholar]
- 25.Mimori T, Hardin J A, Steitz J A. J Biol Chem. 1986;261:2274–2278. [PubMed] [Google Scholar]
- 26.Reeves W H, Sthoeger Z M. J Biol Chem. 1989;264:5047–5052. [PubMed] [Google Scholar]
- 27.Smider V, Rathmell W K, Lieber M R, Chu G. Science. 1994;266:288–291. doi: 10.1126/science.7939667. [DOI] [PubMed] [Google Scholar]
- 28.Boubnov N V, Hall K T, Will Z, Lee S E, He D M, Benjamin D M, Pilaski C R, Band H, Reeves W, Hendrickson E A, Weaver D T. Proc Natl Acad Sci USA. 1995;92:890–894. doi: 10.1073/pnas.92.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizuta R, Taccioli G E, Alt F W. Int Immunol. 1996;8:1467–1471. doi: 10.1093/intimm/8.9.1467. [DOI] [PubMed] [Google Scholar]
- 30.Errami A, Smider V, Rathmell W K, He D M, Hendrickson E A, Zdzienicka M Z, Chu G. Mol Cell Biol. 1996;16:1516–1526. doi: 10.1128/mcb.16.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singleton B K, Priestley A, Steingrimsdottir H, Gell D, Blunt T, Jackson S P, Lehmann R, Jeggo P A. Mol Cell Biol. 1997;17:1264–1273. doi: 10.1128/mcb.17.3.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nussenzweig A, Chen C, da Costa Soares V, Sanchez M, Sokol K, Nussenzweig M C, Li G C. Nature (London) 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- 33.Zhu C, Bogue M A, Lim D-S, Hasty P, Roth D B. Cell. 1996;86:379–389. doi: 10.1016/s0092-8674(00)80111-7. [DOI] [PubMed] [Google Scholar]
- 34.Gottlieb T M, Jackson S P. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 35.Finnie N J, Gottlieb T M, Blunt T, Jeggo P A, Jackson S P. Proc Natl Acad Sci USA. 1995;92:320–324. doi: 10.1073/pnas.92.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peterson S R, Kurimasa A, Oshimura M, Dynan W S, Bradbury E M, Chen D J. Proc Natl Acad Sci USA. 1995;92:3171–3174. doi: 10.1073/pnas.92.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirchgessner C U, Patil C K, Evans J W, Cuomo C A, Fried L M, Carter T, Oettinger M A, Brown J M. Science. 1995;267:1178–1183. doi: 10.1126/science.7855601. [DOI] [PubMed] [Google Scholar]
- 38.Sipley J D, Menninger J C, Hartley K O, Ward D C, Jackson S P, Anderson C W. Proc Natl Acad Sci USA. 1995;92:7515–7519. doi: 10.1073/pnas.92.16.7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller R D, Hogg J, Ozaki J H, Gell D, Jackson S P, Riblet R. Proc Natl Acad Sci USA. 1995;92:10792–10795. doi: 10.1073/pnas.92.23.10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blunt T, Gell D, Fox M, Taccioli G E, Lehmann A R, Jackson S P, Jeggo P A. Proc Natl Acad Sci USA. 1996;93:10285–10290. doi: 10.1073/pnas.93.19.10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Danska J S, Holland D P, Mariathasan S, Williams K M, Guidos C J. Mol Cell Biol. 1996;16:5507–5517. doi: 10.1128/mcb.16.10.5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson L H, Jeggo P A. Mutat Res. 1995;337:131–4. doi: 10.1016/0921-8777(95)00018-f. [DOI] [PubMed] [Google Scholar]
- 43.Li Z, Otevrel T, Gao Y, Cheng H-L, Seed B, Stamato T, Taccioli G E, Alt F W. Cell. 1995;83:1079–1089. doi: 10.1016/0092-8674(95)90135-3. [DOI] [PubMed] [Google Scholar]
- 44.Lee S E, He D M, Hendrickson E A. In: Molecular Analysis of DNA Rearrangements in the Immune System. Jessberger R, Lieber M R, editors. Berlin: Springer; 1996. pp. 133–142. [Google Scholar]
- 45.Gorman J R, van der Stoep N, Monroe R, Cogne M, Davidson L, Alt F W. Immunity. 1996;5:241–252. doi: 10.1016/s1074-7613(00)80319-2. [DOI] [PubMed] [Google Scholar]
- 46.Li E, Bestor T H, Jaenisch R. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 47.Kuhn R, Rajewsky K, Muller W. Science. 1991;254:707–710. doi: 10.1126/science.1948049. [DOI] [PubMed] [Google Scholar]
- 48.Yenofsky R L, Fine M, Pellow J W. Proc Natl Acad Sci USA. 1990;87:3435–3439. doi: 10.1073/pnas.87.9.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hesse J E, Lieber M R, Gellert M, Mizuuchi K. Cell. 1987;49:775–783. doi: 10.1016/0092-8674(87)90615-5. [DOI] [PubMed] [Google Scholar]
- 50.Takiguchi Y, Kurimasa A, Chen F, Pardington P E, Kyama T, Okinaka R T, Moyzis R, Chen D J. Genomics. 1996;35:129–135. doi: 10.1006/geno.1996.0331. [DOI] [PubMed] [Google Scholar]
- 51.Griffith A J, Craft J, Evens J, Mimori T, Hardin J A. Mol Biol Rep. 1992;16:91–97. doi: 10.1007/BF00419754. [DOI] [PubMed] [Google Scholar]
- 52.Anderson C W, Carter T H. In: Molecular Analysis of DNA Rearrangements in the Immune System. Jessberger R, Lieber M R, editors. Berlin: Springer; 1996. pp. 91–111. [Google Scholar]
- 53.Cao Q, Pitt S, Leszyk J, Baril E. Biochemistry. 1994;33:8548–8557. doi: 10.1021/bi00194a021. [DOI] [PubMed] [Google Scholar]
- 54.Tuteja N, Tuteja R, Ochem A, Taneja P, Huang N, Simoncsits A, Susic S, Rahman K, Marusic L, Chen J, Zhang J, Wang S, Pongor S, Falaschi A. EMBO J. 1994;13:4991–5001. doi: 10.1002/j.1460-2075.1994.tb06826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giffin W, Torrance H, Rodda D J, Prefontaine G G, Pope L, Hache R J G. Nature (London) 1996;380:265–268. doi: 10.1038/380265a0. [DOI] [PubMed] [Google Scholar]
- 56.Malynn B A, Blackwell T K, Fulop G M, Rathbun G A, Furley A J W, Ferrier P, Heinke L B, Phillips R A, Yancopoulos G D, Alt F W. Cell. 1988;54:453–460. doi: 10.1016/0092-8674(88)90066-9. [DOI] [PubMed] [Google Scholar]
- 57.Lieber M R, Hesse J E, Lewis S, Bosma G C, Mizuuchi K, Bosma M J, Gellert M. Cell. 1988;55:7–16. doi: 10.1016/0092-8674(88)90004-9. [DOI] [PubMed] [Google Scholar]
- 58.Blackwell T K, Malynn B A, Pollock R R, Ferrier P, Covey L R, Fulop G M, Phillips R A, Yancopoulos G D, Alt F W. EMBO J. 1989;8:735–742. doi: 10.1002/j.1460-2075.1989.tb03433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hendrickson E A, Schlissel M S, Weaver D T. Mol Cell Biol. 1990;10:5397–5407. doi: 10.1128/mcb.10.10.5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bosma M J. Immunodefic Rev. 1992;3:261–276. [PubMed] [Google Scholar]