Abstract
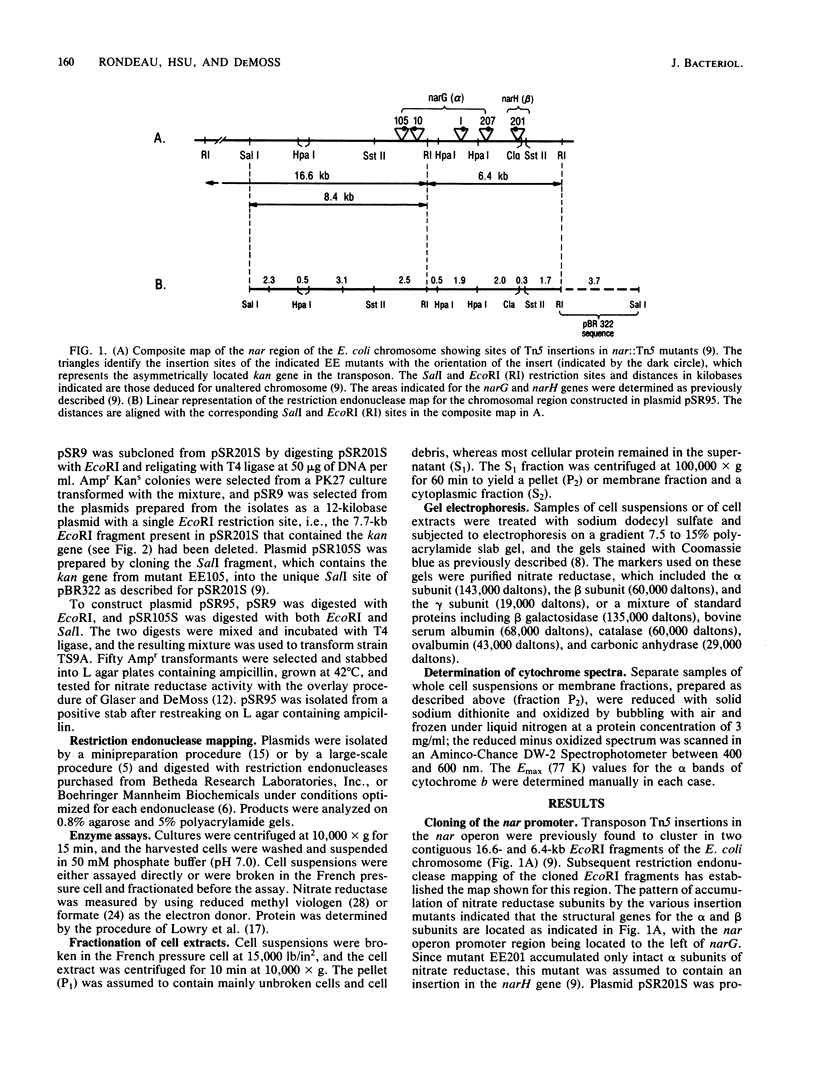
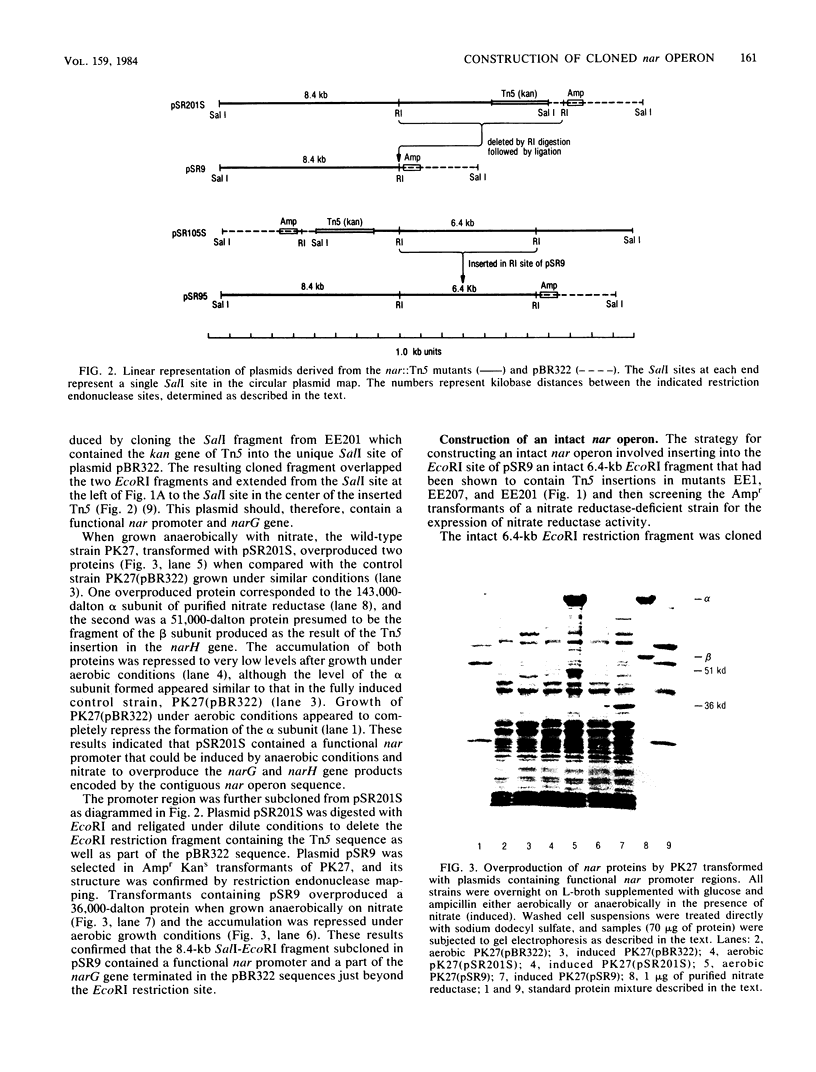
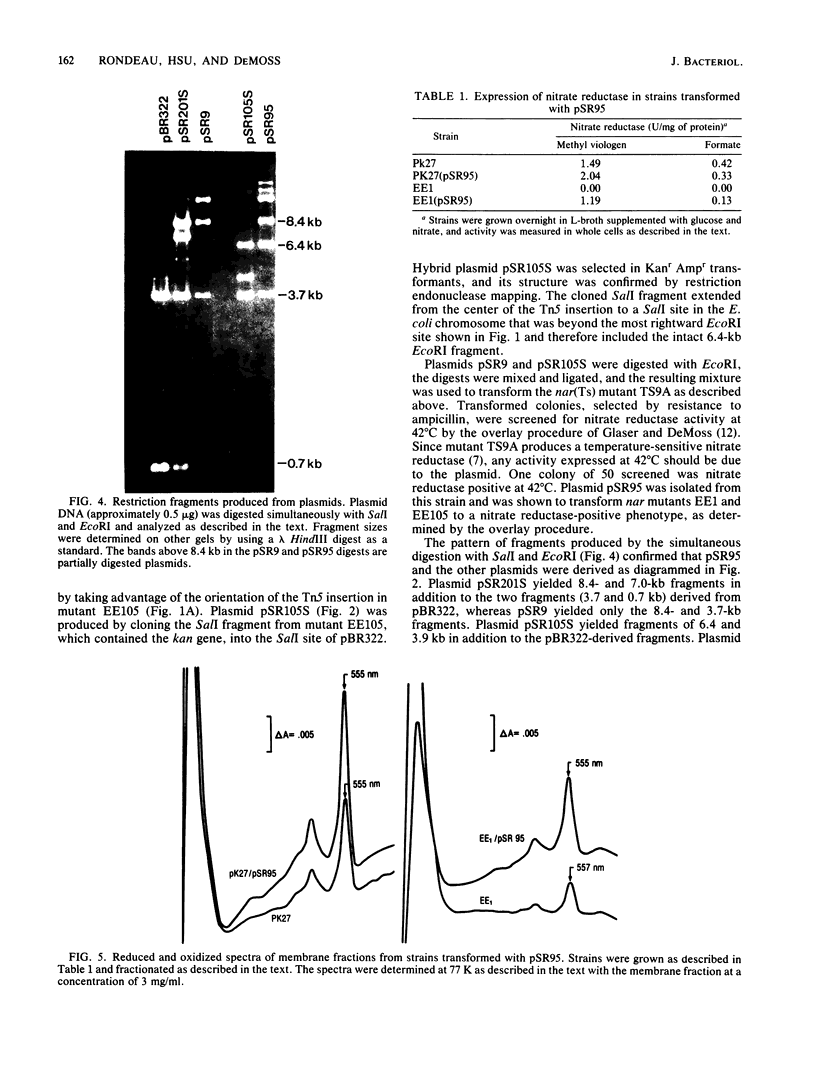
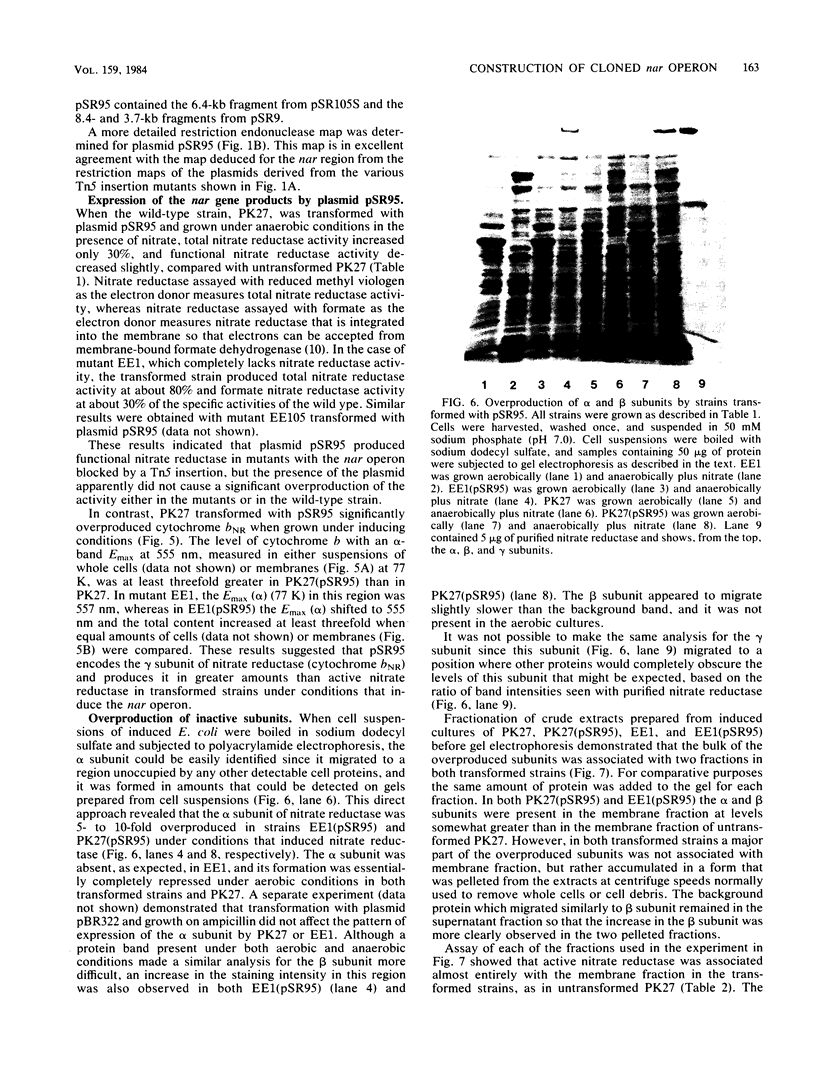
To clone the nar operon of Escherichia coli without an effective selection procedure for the nar+ phenotype, a strategy utilizing nar::Tn5 mutants was employed. Partial segments of the nar operon containing Tn5 insertions were cloned into plasmid pBR322 by using the transposon resistance character for selection. A hybrid plasmid was constructed in vitro from two of these plasmids and isolated by a procedure that involved screening a population of transformed nar(Ts) mutant TS9A for expression of thermal stable nitrate reductase activity. A detailed restriction site map of the resulting plasmid, pSR95, corresponded closely to the composite restriction endonuclease map deduced for the nar region from maps of the cloned nar::Tn5 fragments. When transformed with pSR95, wild-type strain PK27 overproduced the alpha, beta, and gamma subunits of nitrate reductase, although nitrate reductase activity was only slightly increased. The alpha and beta subunits were overproduced about 5- to 10-fold and accumulated mostly as an inactive aggregate in the cytoplasm; the gamma subunit overproduction was detected as a threefold increase in the specific content of cytochrome b555 in the membrane fraction. Functional nitrate reductase and the cytochrome spectrum associated with functional nitrate reductase were restored in the nar::Tn5 mutant EE1 after transformation with pSR95. Although the specific activity of nitrate reductase in this case was less than that of the wild type, both the alpha and beta subunits appeared to be overproduced in an inactive form. In both strains PK27(pSR95) and EE1(pSR95), the formation of nitrate reductase activity and the accumulation of inactive subunits were repressed during aerobic growth. From these observations and the accumulation of inactive subunits were repressed during aerobic growth. From these observations and the demonstration that pSR95 contains a functional nor operon that encodes the alpha, beta, gamma subunits of nitrate reductase.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amy N. K. Identification of the molybdenum cofactor in chlorate-resistant mutants of Escherichia coli. J Bacteriol. 1981 Oct;148(1):274–282. doi: 10.1128/jb.148.1.274-282.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belas R., Mileham A., Cohn D., Hilman M., Simon M., Silverman M. Bacterial bioluminescence: isolation and expression of the luciferase genes from Vibrio harveyi. Science. 1982 Nov 19;218(4574):791–793. doi: 10.1126/science.10636771. [DOI] [PubMed] [Google Scholar]
- Bonnefoy-Orth V., Lepelletier M., Pascal M. C., Chippaux M. Nitrate reductase and cytochrome bnitrate reductase structural genes as parts of the nitrate reductase operon. Mol Gen Genet. 1981;181(4):535–540. doi: 10.1007/BF00428749. [DOI] [PubMed] [Google Scholar]
- Chaudhry G. R., MacGregor C. H. Escherichia coli nitrate reductase subunit A: its role as the catalytic site and evidence for its modification. J Bacteriol. 1983 Apr;154(1):387–394. doi: 10.1128/jb.154.1.387-394.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMoss J. A. Role of the chlC gene in formation of the formate-nitrate reductase pathway in Escherichia coli. J Bacteriol. 1978 Feb;133(2):626–630. doi: 10.1128/jb.133.2.626-630.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demoss J. A., Fan T. Y., Scott R. H. Characterization of subunit structural alterations which occur during purification of nitrate reductase from Escherichia coli. Arch Biochem Biophys. 1981 Jan;206(1):54–64. doi: 10.1016/0003-9861(81)90065-5. [DOI] [PubMed] [Google Scholar]
- Edwards E. S., Rondeau S. S., DeMoss J. A. chlC (nar) operon of Escherichia coli includes structural genes for alpha and beta subunits of nitrate reductase. J Bacteriol. 1983 Mar;153(3):1513–1520. doi: 10.1128/jb.153.3.1513-1520.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch H. G., Lester R. L. The purification and properties of formate dehydrogenase and nitrate reductase from Escherichia coli. J Biol Chem. 1975 Sep 10;250(17):6693–6705. [PubMed] [Google Scholar]
- Glaser J. H., DeMoss J. A. Comparison of nitrate reductase mutants of Escherichia coli selected by alternative procedures. Mol Gen Genet. 1972;116(1):1–10. doi: 10.1007/BF00334254. [DOI] [PubMed] [Google Scholar]
- Glaser J. H., DeMoss J. A. Phenotypic restoration by molybdate of nitrate reductase activity in chlD mutants of Escherichia coli. J Bacteriol. 1971 Nov;108(2):854–860. doi: 10.1128/jb.108.2.854-860.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett N. R., Bragg P. D. Membrane cytochromes of Escherichia coli chl mutants. J Bacteriol. 1983 May;154(2):719–727. doi: 10.1128/jb.154.2.719-727.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N., Roth J., Botstein D. Genetic engineering in vivo using translocatable drug-resistance elements. New methods in bacterial genetics. J Mol Biol. 1977 Oct 15;116(1):125–159. doi: 10.1016/0022-2836(77)90123-1. [DOI] [PubMed] [Google Scholar]
- Klein R. D., Selsing E., Wells R. D. A rapid microscale technique for isolation of recombinant plasmid DNA suitable for restriction enzyme analysis. Plasmid. 1980 Jan;3(1):88–91. doi: 10.1016/s0147-619x(80)90037-2. [DOI] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lund K., DeMoss J. A. Association-dissociation behavior and subunit structure of heat-released nitrate reductase from Escherichia coli. J Biol Chem. 1976 Apr 25;251(8):2207–2216. [PubMed] [Google Scholar]
- MacGregor C. H. Anaerobic cytochrome b1 in Escherichia coli: association with and regulation of nitrate reductase. J Bacteriol. 1975 Mar;121(3):1111–1116. doi: 10.1128/jb.121.3.1111-1116.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor C. H. Biosynthesis of membrane-bound nitrate reductase in Escherichia coli: evidence for a soluble precursor. J Bacteriol. 1976 Apr;126(1):122–131. doi: 10.1128/jb.126.1.122-131.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor C. H., Schnaitman C. A., Normansell D. E. Purification and properties of nitrate reductase from Escherichia coli K12. J Biol Chem. 1974 Aug 25;249(16):5321–5327. [PubMed] [Google Scholar]
- MacGregor C. H., Schnaitman C. A. Reconstitution of nitrate reductase activity and formation of membrane particles from cytoplasmic extracts of chlorate-resistant mutants of Escherichia coli. J Bacteriol. 1973 Jun;114(3):1164–1176. doi: 10.1128/jb.114.3.1164-1176.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Herrera J., DeMoss J. A. Nitrate reductase complex of Escherichia coli K-12: participation of specific formate dehydrogenase and cytochrome b1 components in nitrate reduction. J Bacteriol. 1969 Sep;99(3):720–729. doi: 10.1128/jb.99.3.720-729.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Herrera J., Showe M. K., DeMoss J. A. Nitrate reductase complex of Escherichia coli K-12: isolation and characterization of mutants unable to reduce nitrate. J Bacteriol. 1969 Mar;97(3):1291–1297. doi: 10.1128/jb.97.3.1291-1297.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R. H., DeMoss J. A. Formation of the formate-nitrate electron transport pathway from inactive components in Escherichia coli. J Bacteriol. 1976 Apr;126(1):478–486. doi: 10.1128/jb.126.1.478-486.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showe M. K., DeMoss J. A. Localization and regulation of synthesis of nitrate reductase in Escherichia coli. J Bacteriol. 1968 Apr;95(4):1305–1313. doi: 10.1128/jb.95.4.1305-1313.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperl G. T., DeMoss J. A. chlD gene function in molybdate activation of nitrate reductase. J Bacteriol. 1975 Jun;122(3):1230–1238. doi: 10.1128/jb.122.3.1230-1238.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V., MacGregor C. H. Nitrate reductase in Escherichia coli K-12: involvement of chlC, chlE, and chlG loci. J Bacteriol. 1982 Aug;151(2):788–799. doi: 10.1128/jb.151.2.788-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]