Abstract
In mammals, tissue-specific sets of pattern-recognition molecules, including Nod-like receptors (NLR), enable concomitant and sequential detection of microbial-associated molecular patterns from both the extracellular and intracellular microenvironment. Repressing and de-repressing the cytosolic surveillance machinery contributes to vital immune homeostasis and protective responses within specific tissues. Conversely, defective biology of NLR drives the development of recurrent infectious, autoimmune and/or inflammatory diseases by failing to mount barrier functions against pathogens, to tolerate commensals, and/or to instruct the adaptive immune response against microbes. Better decoding microbial strategies that are evolved to circumvent NLR sensing will provide clues for the development of rational therapies aimed at curing and/or preventing common and emerging immunopathologies.
Introduction
Mammals face life-threatening signals and have the double-edged challenge of eliminating infectious agents and tolerating their flora, especially in the gastrointestinal tract. In mammals, the combination of germ-line encoded pattern-recognition molecules (PRM), including Toll-like receptors (TLR) and Nod-like receptors (NLR), plays an essential role in detecting a diversified set of extracellular and intracellular “danger” signals that primarily originate from microbes (so-called microbial-associated molecular patterns [MAMP]) [1,2]. MAMP are highly conserved microbial-derived molecules shared by both pathogens (in which they are designated as PAMP [pathogen-associated molecular patterns]) and commensals, such as lipopolysaccharides, carbohydrates (including peptidoglycans [PGN]), flagellins, nucleic acids, and peptidic and lipidic structures [3]. Originally identified in the fruit fly and plants, the membrane-bound receptors TLR sense MAMP through their extracellular domain, whereas NLR detect signals inside the cells. Upon recognition of their specific MAMP (Table 1), NLR drive innate and adaptive responses and participate in homeostasis within various host tissues through the activation of transcription factors and downstream effectors, such as mitogen-activated protein kinase (MAPK) (Figure 1) [4–9]. Recent studies emphasized major contributions of NLR in microbial pathogenesis and mammalian immunity. Herein, we summarize the mechanisms of microbial detection by NLR, the NLR-mediated immune signaling, the crosstalk between NLR–NLR and NLR–TLR in mammals, and the strategies used by pathogens to circumvent NLR signaling. Lastly, we will discuss the pathophysiological implications of both NLR and TLR in human diseases, because mutations in NLR- and TLR-encoding genes have been linked to chronic inflammatory diseases, resistance and susceptibility toward a panel of infectious agents, and/or autoimmunity.
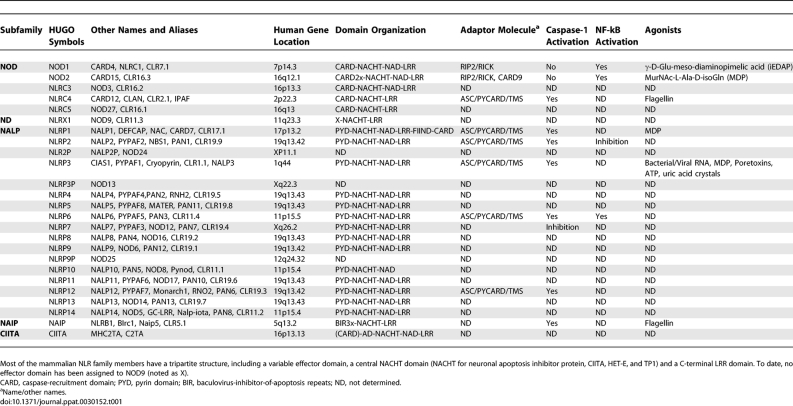
Table 1.
NLR, MAMP, and “Danger” Signals
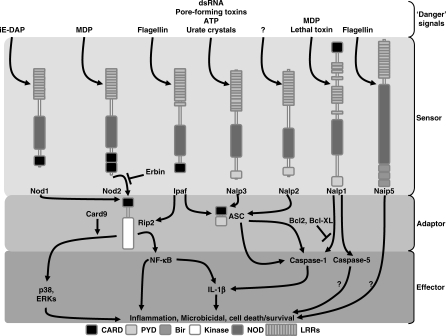
Figure 1. Intracellular Debugging of the NLR Signaling Pathways.
A schematic overview of the major NLR signaling pathways in innate immunity is depicted. Upon detection of their agonists, NLR likely oligomerize through the NOD domain and recruit at least three specific adaptors, including RIP2, CARD9, and ASC. Several modulators of NLR signaling have been recently identified, such as Erbin, Bcl2, and Bcl-xl. The maturation of IL-1β by the inflammasome illustrates the interplay between NLR (i.e., NALP1–3, NAIP5, and IPAF) and other PRM, such as TLR. Better understanding of the spatio-temporal engagement and/or repression of specific NLR might open new avenues for therapeutical intervention.
NLR Are Cytosolic Biosensors for Both Intra- and Extracellular Microbes
Similarly to the superfamily of plant disease-resistance proteins [10], the structure of NLR, also referred as caterpillers, is composed of a N-terminal effector domain, a central oligomerization domain (called NACHT for neuronal apoptosis inhibitor protein, CIITA, HET-E, and TP1), and a C-terminal collection of leucine-rich repeats (LRR) [4–7]. A set of about 23 mammalian NLR has been recently identified by in silico mining of genomic databases for proteins with homology to the apoptosis regulator Apaf-1. NLR are classified accordingly to their effector domains, caspase-recruitment domain (CARD) for nucleotide-binding oligomerization domain protein (NOD), the pyrin domain (PYD) for NALP, and the baculovirus-inhibitor-of-apoptosis repeats (BIR) for NAIP (Figure 1 and Table 1). The effector modules CARD and PYD belong to the death-domain family and define the specificity of the cellular response by activating multiple signaling pathways through homophilic and heterophilic protein-protein association.
The NOD-dependent signaling pathway.
Bacterial PGN is a parietal structure found in all proteobacteria, and both NOD1 and NOD2 have been identified as sensors of fragments derived from PGN, namely muramyl dipeptide (MDP) for NOD2 [11–13] and γ-D-Glu-meso-DAP (iE-DAP) for NOD1 [14,15]. Upon recognition of MDP or iE-DAP, the CARD-containing serine/threonine kinase RIP2 (also known as RICK, CARDIAK, CCK, and Ripk2) engages inflammatory and antimicrobial responses independently of TLR [16–18]. The NOD1- and NOD2-dependent immune response can be triggered by invasive bacteria that replicate inside cells, such as Listeria monocytogenes [16,17,19] (Figure 1). Whereas RIP2-deficient mice have increased susceptibility to systemic infection by L. monocytogenes [18], NOD2-deficient mice showed susceptibility to oral (but not systemic) listeriosis [13]. Extracellular bacterial pathogens can also be sensed by NOD1 and NOD2 through the intracellular delivery of muropeptides by either their type III or IV secretion apparatus [20,21]. These secretion machines are molecular syringes that form transport channels through the membrane of target cells to deliver virulence factors into cytosol. However, the mechanisms by which muropeptides are injected intracellularly remain elusive. Notably, NOD1 is required for the innate immune response to Helicobacter pylori, a major agent of gastric ulcer [20]. Conversely, bacterial mutants deficient in PGN synthesis or type IV secretion system have impaired ability to trigger NOD1-mediated responses in epithelial cells of the stomach [20,22].
More recently, CARD9 has been identified to physically interact with NOD2 and RIP2 to selectively synergize the MDP-induced activation of MAPK but not of nuclear factor–κ B (NF-κB) (Figure 1), providing a mechanism by which CARD9 might control innate immune response toward intracellular pathogens [23]. CARD9 is a CARD-containing adaptor which modulates the development of T(H)-17 response [24] and lacks the C-terminal, membrane-associated guanylate kinase domain. Similarly to RIP2-deficient mice, CARD9-deficient mice are susceptible to systemic infections by L. monocytogenes [23]. Because CARD9 is involved in Dectin1-mediated response to fungal infection [25], the physiological role of NOD1 and NOD2 in response to fungi is now eagerly awaited. Lastly, given that CARD9 is also required for TLR3- and TLR7-dependent signaling pathways [23], further work is now warranted to assess the physiological role of CARD9 in immunity to viruses.
The inflammasome.
The inflammasome is an inflammatory caspase-activating complex, which contain at least caspase-1 and -5, ASC, NALP1-3, IPAF, and NAIP5 [26]. Notably, caspase-5 is recruited by NALP1, but not NALP2 and NALP3, through homophilic CARD–CARD interactions (Figure 1) [26,27]. However, the physiological role of NALP2 and caspase-5 remains poorly understood [28]. Caspase-1, also known as IL-1–converting enzyme (ICE), is a protease involved in pyroptosis, a recently described form of inflammatory programmed cell death, and is essential for the processing of immature pro-inflammatory cytokine IL-1β and the related members IL-1α, IL-18, and IL-33 [29]. The IL-1β is instrumental to initiate and/or amplify innate and adaptive immunity toward pathogens [28]. Caspase-1 activation is also involved in the cleavage of the MyD88-like adaptor Mal [30] and in membrane biogenesis by promoting cell survival following toxin-induced membrane permeabilization [31]. The contribution of caspase-1 and NALP/NAIP in host–pathogen interaction has been analyzed in vivo by using animal models of infection or in vitro by using cytokine and viability assays with monocytes/macrophages. Caspase-1−/− mice, which are inoculated nasally by the agent of bacillary dysentery Shigella flexneri cannot trigger IL-1β–dependent acute inflammation, thereby resulting in exacerbated infection [32]. Interestingly, a positive correlation was defined for S. Typhimurium and S. flexneri between their capacity to promote macrophage death and their virulence in mice. This effect has been proposed to depend on the inflammasome [26,33].
Sensing of several extracellular and intracellular pathogenic bacteria is associated to the secretion of IL-1β by activating caspase-1 through at least five NLR, namely ICE protease-activating factor (IPAF), NALP-1, −2 and −3, and NAIP5 (Figure 1 and Table 1). Containing a N-terminal PYD and a C-terminal CARD, the death-fold–containing adaptor apoptosis-associated speck-like protein containing a CARD (ASC) acts as a molecular bridge between NALP1-3, IPAF, and caspase-1 [26] (Figure 1). In vivo, NALP1 senses the Bacillus anthracis lethal toxin, which is delivered in the cytoplasm by receptor-mediated endocytosis [34]. The bacterial PGN component MDP activates both the NALP1- and NALP3-dependent inflammasome [35,36]. NALP3 is also able to detect a large variety of additional microbial signals (such as microbial RNA) and cytolytic toxins (such as aerolysin from Aeromonas hydrophila and maitotoxin) [31,37–39]. The NALP3-dependent inflammasome is also activated by cellular components that are released into the extracellular milieu by distressed cells such as crystals found in gout, namely monosodium urate and calcium pyrophosphate dihydrate [40]. Oral infection with S. Typhimurium of caspase-1–deficient mice, but not ASC−/−, NALP3−/−, or IPAF−/− animals, leads to increased susceptibility to infection [41]. Similar observations have been reported with L. monocytogenes [42,43]. Furthermore, caspase-1– and ASC-deficient (but not IPAF- and NOD2-deficient) mice experienced increased susceptibility to Francisella tularensis, the agent of tularemia [44].
By using microinjection or liposome delivery or bacterial mutants, the caspase-1 activators, such as IpaB (from S. flexneri), SipB (from S. Typhimurium) and flagellin (from S. Typhimurium and Legionella pneumophila), have been recently proposed to signal through NAIP5 and IPAF [45–50]. Interestingly, NAIP5 is required to limit the maturation of phagosomes following in vitro infection by L. pneumophila, the agent of Legionnaires disease, [51] and restrict its intracellular replication independently of caspase-1 activation [49,52]. IPAF has similar functions that depend, however, on caspase-1 [52,53]. Notably, IPAF and caspase-1, but not ASC, are required to control pyroptosis and autophagy induced by Shigella independently of flagellin [54]. Whereas NAIP5 confers resistance to Legionnaires disease [55–57], the physiological role of IPAF remains elusive with respect to L. pneumophila. Unlike the inflammasome, the sensing of flagellin through TLR5 does not trigger caspase-1 activation, suggesting that IPAF and NAIP5 represent a fail-safe immune mechanism to respond to flagellated pathogens [48,57,58]. Lastly, interferon-β and tumor necrosis factor-α have been implicated in restricting the growth of L. pneumophila in macrophages, but independently of IPAF and NAIP5 [49,59]. Taken together, deficiency in NAIP5, ASC, and/or caspase-1 in mice are, instead, associated with increased susceptibility to invasive microbes and with resistance to the lethal effect of endotoxins [50,60]. Therefore, the NLR signaling pathway and the inflammasome represent “watchdog” machineries against both intra- and extracellular pathogens, including toxicogenic microbes.
Optimal Immune Response Toward Pathogens Requires NLR
Given that several sensors are likely solicited when host cell faces a microorganism, the engagement of specific combinations of NLR and other PRM affect the host response by driving either tolerance, priming, or synergy. Notably, by using cell-based assays, it was found that chemically synthesized NOD1 and NOD2 agonists synergize TLR-dependent cytokines expression in monocytes [61–66]. Similar findings have been reported in dendritic cells (DCs) for the secretion of IL-12 [67,68], which is a major cytokine produced by DCs to promote T(H)1 polarization of T cells [69]. DCs are the central “professional” immune cells for the initiation of the adaptive immunity by activating the T lymphocyte differentiation into T(H)1 or T(H)2 cells. It is worth noting that the RIP2 signaling pathway is required for efficient elicitation of antigen-specific T and B cell immunity and for instructing subsequently the onset of T(H)1 and T(H)2, as well as T(H)17 immune pathways by regulating IL-23, IL-12, interferon-γ and IL-17 [13,18,70]. Conversely, NOD1- and NOD2-deficient bone marrow–derived macrophages and macrophages bearing loss-of-function of NOD2 failed to synergize the expression of inflammatory cytokines following concomitant stimulation by NOD1/2 and TLR agonists [13,14,63]. The role of the inflammasome in the development of autoimmune diseases and T(H)17 response to pathogens and “danger” signals remains to be further documented, because the IL-1 receptor is required to drive the development of experimental autoimmune encephalomyelitis and the production of IL-17 [71,72].
Triggering of a specific PRM induces a transient phenomenon of tolerance and/or cross-tolerance toward a second stimulation by the same agonist [73]. This unresponsiveness window may be essential to protect the host from sustained innate response. If a microorganism colonizes a TLR-responsive niche, one can expect that the TLR signaling might be exhausted. This TLR refractory state is nevertheless specific for this niche and for a certain period of time. In this context, subsequent signaling is likely to occur for unrepressed NLR signaling pathways [74,75]. Alternatively, sequential engagement of distinct TLR stimulates mainly the synergistic production of pro-inflammatory mediators [73]. Therefore, systematic dissection of the synergy and tolerance induced by TLR and/or NLR is now warranted for rationale therapeutic intervention.
The heterotypic interactions between NLR have also been proposed to regulate their function [76,77], as well as interplay with additional proteins like Erbin for the NOD2 signaling pathway [78]. Effective NALP1- and NALP3-dependent inflammasome activation requires both the synthesis of pro-IL-1β and several incoming signals. Activation of TLR signaling pathways has often been used as a first signal that drives the expression of the IL-1β–encoding gene [47–49,58,79–81]. Activation of caspase-1 and release of mature IL-1β might then result from the cytosolic detection by the inflammasome of a second signal (which are primarily microbial compounds and self-danger signals). Alternatively, the intracellular concentration of potassium and ATP/dATP, which might be modulated by toxins and pathogens, control inflammasome activation [80,82–84]. In this context, the transmembrane receptor Pannexin-1 and the P2X(7) receptor participate in the activation of the inflammasome by transferring bacterial components from outside to the cytosol [80]. It is worth noting the increased viability of “permissive” macrophages to replication of L. pneumophila, which is in part explained by a specific targeting of pro-death members of the Bcl2 protein family [85]. Similarly to what occurs in the nematode Caenorhabditis elegans, a complex interplay between cell survival and caspase-1 activation has been recently unraveled, as the mammalian anti-apoptotic proteins Bcl-2 and Bcl-Xl repress specifically the NALP1-mediated caspase-1 activation [35]. Implications for the pathophysiology of human inflammatory diseases and the pathogenesis of pathogens that activate caspase-1, like L. pneumophila, remain to be investigated for the development of rational prophylactic and anti-infectious therapies.
NLR in Antimicrobial Immunity
Antimicrobial peptides, such as defensins, play an active role in fortifying the lining of the gut toward pathogens and commensals by preserving epithelium integrity and stem cell viability and by participating in the recruitment of immunocytes. Defensins are cationic antimicrobial peptides rich in cysteine residues that exert their activity by damaging the bacterial cell wall [86]. Mammalian defensins can be divided into two main subsets, the α and β defensins. In humans, α defensins 1–4 are produced by neutrophils and are stored in granules, whereas α defensins 5 and 6 (referred as cryptdins in mice) are mostly secreted by Paneth cells in the lumen of the small intestine. The human β defensins are more widely expressed and are synthesized by most of the epithelia. Proteases (e.g., trypsin and metalloproteinase-7) play an essential role in the maturation process of defensins [87,88]. Alternatively, the expression of certain antimicrobial peptide-encoding genes is down-regulated during shigellosis in humans and salmonellosis in mice [89,90]. In this context, we might speculate that specific combinations of PRM coordinate a three-steps immune response by regulating expression, degranulation, and maturation of antimicrobial peptides.
Recent evidences shed also lights on a nonredundant role of NLR and TLR in the differential expression and maturation of specific sets of antimicrobial peptides [91]. By using cell lines, NOD1 and NOD2 agonists have been shown to up-regulate the expression of the human β defensin-2 (hBD2)–encoding gene [91]. Likewise, inhibition of the NF-κB pathway totally blocks NOD1- and NOD2-dependent induction of hBD2 expression, whereas inhibition of p38 and JNK signaling pathways only partially diminish hBD2 expression in a NOD1-dependent manner [22,92]. Consistently, MDP-induced hBD2 expression was down-regulated after knocking down NOD2 or by transfecting HEK293 cells with the Crohn disease–associated 3020insC frameshift mutation [92]. In vivo, TLR5-deficient mice exhibit impaired expression of mouse β defensin-3 (the homolog of the hBD-2) by intestinal epithelial cells [93], whereas NOD1- and NOD2-deficient mice showed a deficiency in mouse β defensin-4 by gastric epithelial cells and in certain cryptdins by Paneth cells, respectively [13,22]. Combining experiments using mice and human models might shed light on the potential effect of antimicrobial peptides in response to specific infectious agents and in the control of physiological inflammation.
Microbial Alteration of NLR Sensing and Signaling
Pathogens have also evolved strategies to circumvent their intracellular sensing through NLR such as NOD1 by preventing processing and optimal sensing of their PGN. Indeed, a L. monocytogenes–derived deacetylase, namely PgdA, is required to bypass the early innate immune response of NOD1 by specifically adding N-acetylglucosamine residues to the DAP-type PGN. pgdA mutants triggered sensitivity to the bacteriolytic activity of lysozyme, leading to increased NOD1-dependent interferon-β immune response and have impaired virulence in vivo [94]. Similarly, the PGN hydrolase AmiA is involved in the microbial pathogenesis of H. pylori by limiting the sensing by NOD1 [95]. The implications of post-translational changes of additional MAMP, including MDP, in microbial pathogenesis remain to be systematically investigated.
Commensal and symbiotic microorganisms may actively interfere with NLR-mediated response by down-regulating pro-inflammatory signaling and/or by modifying cell differentiation/proliferation. Bacteroides thetaiotamicron specifically stimulates the nucleus-cytosol shuttling of NF-κB in a peroxisome proliferator–activated receptor-γ–dependent mechanism [96]. The bacterial molecules driving this down-regulation might consequently affect NOD signaling. In addition to commensal, pathogenic bacteria are also able to interfere with NOD, as recently showed in Yersinia species that block stimulation of NF-κB– and MAPK-dependent gene expression through acetylation of MAPK [97]. In other respects, S. flexneri produces a phosphatase that is specific for nuclear MAPK, thus preventing histone H3 modification and NF-κB–dependent transcription of pro-inflammatory genes [98]. Lastly, modification of ubiquitination is also a candidate mechanism for invasive microbes to subvert sensing through NLR and is now the subject of investigation [99]. This conceptual view of regulation of NLR signaling now warrants further investigation to better understand the double-edged challenge of the mucosa toward commensals and pathogens in health and disease.
Impaired TLR and NLR Function Are Sufficient to Drive Human Immunopathologies
Microbial sensing deficiency in fruit fly and mice confers an increased susceptibility to several infectious agents and leads to the development of autoimmune disease. In humans, defective NF-κB– and TLR-dependent immunity have been associated with restricted inherited infectious diseases, in that hypomorphic or null germline mutations of TLR5, IRAK4, NEMO, and UNC-93B are sufficient for the development of Legionnaires disease [100], recurrent pneumococcal disease [101], mycobacterial disease [102], and herpes simplex virus encephalitis [103], respectively. Similarly, common infection might result primarily from mutations in a major susceptibility gene, as impaired TLR2/Mal signaling is conferring protection against invasive pneumococcal disease, bacteremia, malaria, and tuberculosis in the United Kingdom, Vietnam, and several African countries through an increased frequency of the Ser allele at the Mal S180L variant [104]. Hence, these observations have challenged dogmas in the genetics of human infectious diseases, and they might suggest that both rare and common infectious diseases may result from defect in a major biological pathway.
Like infectious diseases, complex inflammatory traits result from the inheritance of major susceptibility alleles and the exposure to several environmental factors. No specific infectious agents have been identified so far, but note that gain-of-function mutations in NALP3 [105] and NOD2 [106] cause Mendelian inflammatory diseases, such as autoinflammatory disorders (Muckle-Wells syndrome, familial cold autoinflammatory syndrome, and chronic infantile neurologic cutaneous and articular syndrome) and Blau syndrome, respectively. Furthermore, predisposition to common inflammatory disorders has been inextricably linked to major susceptibility genes, as loss-of-function mutations in TLR5 and NOD2 are protecting and predisposing, respectively, to the development of systemic lupus erythematosus [107] and inflammatory bowel diseases [108,109]. Similarly, NEMO deficiency in enterocytes leads to the spontaneous development of colitis in mice by compromising tissue repair, epithelium integrity and promoting bacterial translocation [110]. A missense mutation (L155H) and noncoding polymorphism of the NALP1-encoding gene are predisposing to the development of vitiligo-associated multiple autoimmune disease [111], implicating innate immunity in the pathogenesis of autoimmunity. However, the constitutive and bacterial-induced level of caspase-1 activation remains elusive in cells bearing the NALP1 L155H mutation. Lastly, mutations in the gene encoding for NALP7 cause familial and recurrent hydatidiform moles [112–114], which are tumors that forms in the uterus as a mass of cysts resembling a bunch of grapes. Unlike NALP1-3, IPAF, and NAIP5, NALP7 is a negative regulator of IL-1β signaling [115] that promotes tumorigenesis [116]. Further work should now determine the following: (a) how the NALP7 signaling pathway might be activated, (b) whether NALP7 might interfere with additional PRM, and (c) whether NALP7 might regulate innate and adaptive immunity.
Concluding Remarks
Whereas most studies have focused on the characterization of individual PRM, the physiological situation is more complex, because host cells have to integrate multiple incoming signals from damaged cells and pathogenic and symbiotic microbes into vital immunological information. In this context, organ-specific onset of innate and adaptive immunity is regulated differentially by multiple cross-talks between a limited set of functional PRM signaling pathways.
Acknowledgments
We apologize to our colleagues whose work was not cited here owing to space limitations. We thank Florent Sebbane for critical reading of the manuscript.
Abbreviations
- ASC
apoptosis-associated speck-like protein containing a caspase recruitment domain
- CARD
caspase-recruitment domain
- DC
dendritic cell
- ICE
IL-1–converting enzyme
- IPAF
ICE protease-activating factor
- LRR
leucine-rich repeat
- MAMP
microbial-associated molecular patterns
- MAPK
mitogen-activated protein kinase
- MDP
muramyl dipeptide
- MyD88
myeloid differentiation primary response gene (88)
- NF-κB
nuclear factor–κ B
- NLR
Nod-like receptors
- NOD
nucleotide-binding oligomerization domain protein
- PGN
peptidoglycan
- PRM
pattern-recognition molecules
- PYD
pyrin domain
- TLR
Toll-like receptors
Footnotes
Jean-Claude Sirard and Rodrigue Dessein are with INSERM, U801, Institut Pasteur de Lille, Université Lille 2, Lille, France. Cécile Vignal and Mathias Chamaillard are with INSERM, U795, Université Lille 2, Lille, France.
Author contributions. JCS, CV, RD, and MC wrote the manuscript.
Funding. JCS is supported by INSERM, the Institut Pasteur de Lille, the Région Nord Pas de Calais and FEDER (ARCir émergence), the European Community (STREP grant VaccTIP LSHP-CT-2005-012161 SavinMucoPath INCO-CT-2006-032296), and the Franco-Argentinean ECOS-SETCIP program (A04B03). CV is supported by the Fondation pour la Recherche Médicale. MC is supported by grants from INSERM, the Région Nord Pas de Calais, the Association François AuPetit, and the IRMAD.
Competing interests. The authors have declared that no competing interests exist.
References
- Matzinger P. Friendly and dangerous signals: is the tissue in control? Nat Immunol. 2007;8:11–13. doi: 10.1038/ni0107-11. [DOI] [PubMed] [Google Scholar]
- Raz E. Organ-specific regulation of innate immunity. Nat Immunol. 2007;8:3–4. doi: 10.1038/ni0107-3. [DOI] [PubMed] [Google Scholar]
- Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- Tschopp J, Martinon F, Burns K. NALPs: a novel protein family involved in inflammation. Nat Rev Mol Cell Biol. 2003;4:95–104. doi: 10.1038/nrm1019. [DOI] [PubMed] [Google Scholar]
- Ting JP, Davis BK. CATERPILLER: a novel gene family important in immunity, cell death, and diseases. Annu Rev Immunol. 2005;23:387–414. doi: 10.1146/annurev.immunol.23.021704.115616. [DOI] [PubMed] [Google Scholar]
- Inohara N, Chamaillard M, McDonald C, Nunez G. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu Rev Biochem. 2005;74:355–383. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- Fritz JH, Ferrero RL, Philpott DJ, Girardin SE. Nod-like proteins in immunity, inflammation and disease. Nat Immunol. 2006;7:1250–1257. doi: 10.1038/ni1412. [DOI] [PubMed] [Google Scholar]
- Barton GM, Medzhitov R. Toll-like receptor signaling pathways. Science. 2003;300:1524–1525. doi: 10.1126/science.1085536. [DOI] [PubMed] [Google Scholar]
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278:8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J Biol Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4:702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jehanno M, et al. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science. 2003;300:1584–1587. doi: 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- Park JH, Kim YG, McDonald C, Kanneganti TD, Hasegawa M, et al. RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J Immunol. 2007;178:2380–2386. doi: 10.4049/jimmunol.178.4.2380. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Inohara N, Hernandez LD, Galan JE, Nunez G, et al. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature. 2002;416:194–199. doi: 10.1038/416194a. [DOI] [PubMed] [Google Scholar]
- Chin AI, Dempsey PW, Bruhn K, Miller JF, Xu Y, et al. Involvement of receptor-interacting protein 2 in innate and adaptive immune responses. Nature. 2002;416:190–194. doi: 10.1038/416190a. [DOI] [PubMed] [Google Scholar]
- O'Riordan M, Yi CH, Gonzales R, Lee KD, Portnoy DA. Innate recognition of bacteria by a macrophage cytosolic surveillance pathway. Proc Natl Acad Sci U S A. 2002;99:13861–13866. doi: 10.1073/pnas.202476699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, et al. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol. 2004;5:1166–1174. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- Girardin SE, Tournebize R, Mavris M, Page AL, Li X, et al. CARD4/Nod1 mediates NF-kappaB and JNK activation by invasive Shigella flexneri. EMBO Rep. 2001;2:736–742. doi: 10.1093/embo-reports/kve155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boughan PK, Argent RH, Body-Malapel M, Park JH, Ewings KE, et al. Nucleotide-binding oligomerization domain-1 and epidermal growth factor receptor: critical regulators of beta-defensins during Helicobacter pylori infection. J Biol Chem. 2006;281:11637–11648. doi: 10.1074/jbc.M510275200. [DOI] [PubMed] [Google Scholar]
- Hsu YM, Zhang Y, You Y, Wang D, Li H, et al. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nat Immunol. 2007;8:198–205. doi: 10.1038/ni1426. [DOI] [PubMed] [Google Scholar]
- Leibundgut-Landmann S, Gross O, Robinson MJ, Osorio F, Slack EC, et al. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat Immunol. 2007;8:630–638. doi: 10.1038/ni1460. [DOI] [PubMed] [Google Scholar]
- Gross O, Gewies A, Finger K, Schafer M, Sparwasser T, et al. Card9 controls a non-TLR signalling pathway for innate anti-fungal immunity. Nature. 2006;442:651–656. doi: 10.1038/nature04926. [DOI] [PubMed] [Google Scholar]
- Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- Agostini L, Martinon F, Burns K, McDermott MF, Hawkins PN, et al. NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity. 2004;20:319–325. doi: 10.1016/s1074-7613(04)00046-9. [DOI] [PubMed] [Google Scholar]
- Martinon F, Tschopp J. Inflammatory caspases: linking an intracellular innate immune system to autoinflammatory diseases. Cell. 2004;117:561–574. doi: 10.1016/j.cell.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Dinarello CA. The IL-1 family and inflammatory diseases. Clin Exp Rheumatol. 2002;20:S1–S13. [PubMed] [Google Scholar]
- Miggin SM, Palsson-McDermott E, Dunne A, Jefferies C, Pinteaux E, et al. NF-kappaB activation by the Toll-IL-1 receptor domain protein MyD88 adapter-like is regulated by caspase-1. Proc Natl Acad Sci U S A. 2007;104:3372–3377. doi: 10.1073/pnas.0608100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurcel L, Abrami L, Girardin S, Tschopp J, van der Goot FG. Caspase-1 activation of lipid metabolic pathways in response to bacterial pore-forming toxins promotes cell survival. Cell. 2006;126:1135–1145. doi: 10.1016/j.cell.2006.07.033. [DOI] [PubMed] [Google Scholar]
- Sansonetti PJ, Phalipon A, Arondel J, Thirumalai K, Banerjee S, et al. Caspase-1 activation of IL-1beta and IL-18 are essential for Shigella flexneri-induced inflammation. Immunity. 2000;12:581–590. doi: 10.1016/s1074-7613(00)80209-5. [DOI] [PubMed] [Google Scholar]
- Mariathasan S, Monack DM. Inflammasome adaptors and sensors: intracellular regulators of infection and inflammation. Nat Rev Immunol. 2007;7:31–40. doi: 10.1038/nri1997. [DOI] [PubMed] [Google Scholar]
- Boyden ED, Dietrich WF. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet. 2006;38:240–244. doi: 10.1038/ng1724. [DOI] [PubMed] [Google Scholar]
- Bruey JM, Bruey-Sedano N, Luciano F, Zhai D, Balpai R, et al. Bcl-2 and Bcl-XL regulate proinflammatory caspase-1 activation by interaction with NALP1. Cell. 2007;129:45–56. doi: 10.1016/j.cell.2007.01.045. [DOI] [PubMed] [Google Scholar]
- Martinon F, Agostini L, Meylan E, Tschopp J. Identification of bacterial muramyl dipeptide as activator of the NALP3/cryopyrin inflammasome. Curr Biol. 2004;14:1929–1934. doi: 10.1016/j.cub.2004.10.027. [DOI] [PubMed] [Google Scholar]
- Kanneganti TD, Ozoren N, Body-Malapel M, Amer A, Park JH, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440:233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- Kanneganti TD, Body-Malapel M, Amer A, Park JH, Whitfield J, et al. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J Biol Chem. 2006;281:36560–36568. doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- Mariathasan S, Weiss DS, Newton K, McBride J, O'Rourke K, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–32. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:228–232. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- Lara-Tejero M, Sutterwala FS, Ogura Y, Grant EP, Bertin J, et al. Role of the caspase-1 inflammasome in Salmonella typhimurium pathogenesis. J Exp Med. 2006;203:1407–1412. doi: 10.1084/jem.20060206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozoren N, Masumoto J, Franchi L, Kanneganti TD, Body-Malapel M, et al. Distinct roles of TLR2 and the adaptor ASC in IL-1beta/IL-18 secretion in response to Listeria monocytogenes. J Immunol. 2006;176:4337–4342. doi: 10.4049/jimmunol.176.7.4337. [DOI] [PubMed] [Google Scholar]
- Tsuji NM, Tsutsui H, Seki E, Kuida K, Okamura H, et al. Roles of caspase-1 in Listeria infection in mice. Int Immunol. 2004;16:335–343. doi: 10.1093/intimm/dxh041. [DOI] [PubMed] [Google Scholar]
- Mariathasan S, Weiss DS, Dixit VM, Monack DM. Innate immunity against Francisella tularensis is dependent on the ASC/caspase-1 axis. J Exp Med. 2005;202:1043–1049. doi: 10.1084/jem.20050977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersh D, Monack DM, Smith MR, Ghori N, Falkow S, et al. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc Natl Acad Sci U S A. 1999;96:2396–2401. doi: 10.1073/pnas.96.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Smith MR, Thirumalai K, Zychlinsky A. A bacterial invasin induces macrophage apoptosis by binding directly to ICE. Embo J. 1996;15:3853–3860. [PMC free article] [PubMed] [Google Scholar]
- Franchi L, Amer A, Body-Malapel M, Kanneganti TD, Ozoren N, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol. 2006;7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- Miao EA, Alpuche-Aranda CM, Dors M, Clark AE, Bader MW, et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol. 2006;7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- Molofsky AB, Byrne BG, Whitfield NN, Madigan CA, Fuse ET, et al. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J Exp Med. 2006;203:1093–1104. doi: 10.1084/jem.20051659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Newton K, Monack DM, Vucic D, French DM, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- Fortier A, de Chastellier C, Balor S, Gros P. Birc1e/Naip5 rapidly antagonizes modulation of phagosome maturation by Legionella pneumophila. Cell Microbiol. 2007;9:910–923. doi: 10.1111/j.1462-5822.2006.00839.x. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M, Amer A, Kanneganti TD, Munoz-Planillo R, Chen G, et al. The Nod-like receptor family member Naip5/Birc1e restricts Legionella pneumophila growth independently of caspase-1 activation. J Immunol. 2007;178:8022–8027. doi: 10.4049/jimmunol.178.12.8022. [DOI] [PubMed] [Google Scholar]
- Amer A, Franchi L, Kanneganti TD, Body-Malapel M, Ozoren N, et al. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J Biol Chem. 2006;281:35217–35223. doi: 10.1074/jbc.M604933200. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Franchi L, Toma C, Ashida H, Ogawa M, et al. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 2007;3:e111. doi: 10.1371/journal.ppat.0030111. doi: 10.1371/journal.ppat.0030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez E, Lee SH, Gauthier S, Yaraghi Z, Tremblay M, et al. Birc1e is the gene within the Lgn1 locus associated with resistance to Legionella pneumophila. Nat Genet. 2003;33:55–60. doi: 10.1038/ng1065. [DOI] [PubMed] [Google Scholar]
- Wright EK, Goodart SA, Growney JD, Hadinoto V, Endrizzi MG, et al. Naip5 affects host susceptibility to the intracellular pathogen Legionella pneumophila. Curr Biol. 2003;13:27–36. doi: 10.1016/s0960-9822(02)01359-3. [DOI] [PubMed] [Google Scholar]
- Zamboni DS, Kobayashi KS, Kohlsdorf T, Ogura Y, Long EM, et al. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat Immunol. 2006;7:318–325. doi: 10.1038/ni1305. [DOI] [PubMed] [Google Scholar]
- Ren T, Zamboni DS, Roy CR, Dietrich WF, Vance RE. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2006;2:e18. doi: 10.1371/journal.ppat.0020018. doi: 10.1371/journal.ppat.0020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coers J, Vance RE, Fontana MF, Dietrich WF. Restriction of Legionella pneumophila growth in macrophages requires the concerted action of cytokine and Naip5/Ipaf signalling pathways. Cell Microbiol. 2007;9:2344–2357. doi: 10.1111/j.1462-5822.2007.00963.x. [DOI] [PubMed] [Google Scholar]
- Li P, Allen H, Banerjee S, Franklin S, Herzog L, et al. Mice deficient in IL-1 beta-converting enzyme are defective in production of mature IL-1 beta and resistant to endotoxic shock. Cell. 1995;80:401–411. doi: 10.1016/0092-8674(95)90490-5. [DOI] [PubMed] [Google Scholar]
- Takada H, Yokoyama S, Yang S. Enhancement of endotoxin activity by muramyldipeptide. J Endotoxin Res. 2002;8:337–342. doi: 10.1179/096805102125000669. [DOI] [PubMed] [Google Scholar]
- Uehara A, Yang S, Fujimoto Y, Fukase K, Kusumoto S, et al. Muramyldipeptide and diaminopimelic acid-containing desmuramylpeptides in combination with chemically synthesized Toll-like receptor agonists synergistically induced production of interleukin-8 in a NOD2- and NOD1-dependent manner, respectively, in human monocytic cells in culture. Cell Microbiol. 2005;7:53–61. doi: 10.1111/j.1462-5822.2004.00433.x. [DOI] [PubMed] [Google Scholar]
- van Heel DA, Ghosh S, Butler M, Hunt KA, Lundberg AM, et al. Muramyl dipeptide and toll-like receptor sensitivity in NOD2-associated Crohn's disease. Lancet. 2005;365:1794–1796. doi: 10.1016/S0140-6736(05)66582-8. [DOI] [PubMed] [Google Scholar]
- van Heel DA, Ghosh S, Butler M, Hunt K, Foxwell BM, et al. Synergistic enhancement of Toll-like receptor responses by NOD1 activation. Eur J Immunol. 2005;35:2471–2476. doi: 10.1002/eji.200526296. [DOI] [PubMed] [Google Scholar]
- van Heel DA, Ghosh S, Hunt K, Mathew C, Forbes A, et al. Synergy between TLR9 and NOD2 innate immune responses is lost in genetic Crohn's disease. Gut. 2005;54:1553–1537. doi: 10.1136/gut.2005.065888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Tamai R, Akashi S, Takeuchi O, Akira S, et al. Synergistic effect of muramyldipeptide with lipopolysaccharide or lipoteichoic acid to induce inflammatory cytokines in human monocytic cells in culture. Infect Immun. 2001;69:2045–2053. doi: 10.1128/IAI.69.4.2045-2053.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz JH, Girardin SE, Fitting C, Werts C, Mengin-Lecreulx D, et al. Synergistic stimulation of human monocytes and dendritic cells by Toll-like receptor 4 and NOD1- and NOD2-activating agonists. Eur J Immunol. 2005;35:2459–2470. doi: 10.1002/eji.200526286. [DOI] [PubMed] [Google Scholar]
- Tada H, Aiba S, Shibata K, Ohteki T, Takada H. Synergistic effect of Nod1 and Nod2 agonists with toll-like receptor agonists on human dendritic cells to generate interleukin-12 and T helper type 1 cells. Infect Immun. 2005;73:7967–7976. doi: 10.1128/IAI.73.12.7967-7976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol. 2003;3:133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- Fritz JH, Le Bourhis L, Sellge G, Magalhaes JG, Fsihi H, et al. Nod1-mediated innate immune recognition of peptidoglycan contributes to the onset of adaptive immunity. Immunity. 2007;26:445–459. doi: 10.1016/j.immuni.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Schnyder-Candrian S, Togbe D, Couillin I, Mercier I, Brombacher F, et al. Interleukin-17 is a negative regulator of established allergic asthma. J Exp Med. 2006;203:2715–2725. doi: 10.1084/jem.20061401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton C, Brereton C, Keogh B, Mills KH, Lavelle EC. A crucial role for interleukin (IL)-1 in the induction of IL-17-producing T cells that mediate autoimmune encephalomyelitis. J Exp Med. 2006;203:1685–1691. doi: 10.1084/jem.20060285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi A, Herrup EA, Warren HS, Trigilio J, Shin HS, et al. MyD88-dependent and MyD88-independent pathways in synergy, priming, and tolerance between TLR agonists. J Immunol. 2007;178:1164–1171. doi: 10.4049/jimmunol.178.2.1164. [DOI] [PubMed] [Google Scholar]
- Serbina NV, Kuziel W, Flavell R, Akira S, Rollins B, et al. Sequential MyD88-independent and -dependent activation of innate immune responses to intracellular bacterial infection. Immunity. 2003;19:891–901. doi: 10.1016/s1074-7613(03)00330-3. [DOI] [PubMed] [Google Scholar]
- Weiss DS, Raupach B, Takeda K, Akira S, Zychlinsky A. Toll-like receptors are temporally involved in host defense. J Immunol. 2004;172:4463–4469. doi: 10.4049/jimmunol.172.7.4463. [DOI] [PubMed] [Google Scholar]
- Damiano JS, Oliveira V, Welsh K, Reed JC. Heterotypic interactions among NACHT domains: implications for regulation of innate immune responses. Biochem J. 2004;381:213–219. doi: 10.1042/BJ20031506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor A, Martinon F, De Smedt T, Petrilli V, Tschopp J. A crucial function of SGT1 and HSP90 in inflammasome activity links mammalian and plant innate immune responses. Nat Immunol. 2007;8:497–503. doi: 10.1038/ni1459. [DOI] [PubMed] [Google Scholar]
- McDonald C, Chen FF, Ollendorff V, Ogura Y, Marchetto S, et al. A role for Erbin in the regulation of Nod2-dependent NF-kappaB signaling. J Biol Chem. 2005;280:40301–40309. doi: 10.1074/jbc.M508538200. [DOI] [PubMed] [Google Scholar]
- Franchi L, McDonald C, Kanneganti TD, Amer A, Nunez G. Nucleotide-binding oligomerization domain-like receptors: intracellular pattern recognition molecules for pathogen detection and host defense. J Immunol. 2006;177:3507–3513. doi: 10.4049/jimmunol.177.6.3507. [DOI] [PubMed] [Google Scholar]
- Kanneganti TD, Lamkanfi M, Kim YG, Chen G, Park JH, et al. Pannexin-1-mediated recognition of bacterial molecules activates the cryopyrin inflammasome independent of Toll-like receptor signaling. Immunity. 2007;26:433–443. doi: 10.1016/j.immuni.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Sutterwala FS, Ogura Y, Szczepanik M, Lara-Tejero M, Lichtenberger GS, et al. Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity. 2006;24:317–327. doi: 10.1016/j.immuni.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Duncan JA, Bergstralh DT, Wang Y, Willingham SB, Ye Z, et al. Cryopyrin/NALP3 binds ATP/dATP, is an ATPase, and requires ATP binding to mediate inflammatory signaling. Proc Natl Acad Sci U S A. 2007;104:8041–8046. doi: 10.1073/pnas.0611496104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L, Kanneganti TD, Dubyak GR, Nunez G. Differential requirement of P2X7 receptor and intracellular K+ for caspase-1 activation induced by intracellular and extracellular bacteria. J Biol Chem. 2007;282:18810–18818. doi: 10.1074/jbc.M610762200. [DOI] [PubMed] [Google Scholar]
- Petrilli V, Papin S, Dostert C, Mayor A, Martinon F, et al. Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell Death Differ. 2007;14:1583–1589. doi: 10.1038/sj.cdd.4402195. [DOI] [PubMed] [Google Scholar]
- Banga S, Gao P, Shen X, Fiscus V, Zong WX, et al. Legionella pneumophila inhibits macrophage apoptosis by targeting pro-death members of the Bcl2 protein family. Proc Natl Acad Sci U S A. 2007;104:5121–5126. doi: 10.1073/pnas.0611030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- Ghosh D, Porter E, Shen B, Lee SK, Wilk D, et al. Paneth cell trypsin is the processing enzyme for human defensin-5. Nat Immunol. 2002;3:583–590. doi: 10.1038/ni797. [DOI] [PubMed] [Google Scholar]
- Wilson CL, Heppner KJ, Rudolph LA, Matrisian LM. The metalloproteinase matrilysin is preferentially expressed by epithelial cells in a tissue-restricted pattern in the mouse. Mol Biol Cell. 1995;6:851–869. doi: 10.1091/mbc.6.7.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam D, Bandholtz L, Nilsson J, Wigzell H, Christensson B, et al. Downregulation of bactericidal peptides in enteric infections: a novel immune escape mechanism with bacterial DNA as a potential regulator. Nat Med. 2001;7:180–185. doi: 10.1038/84627. [DOI] [PubMed] [Google Scholar]
- Salzman NH, Chou MM, de Jong H, Liu L, Porter EM, et al. Enteric salmonella infection inhibits Paneth cell antimicrobial peptide expression. Infect Immun. 2003;71:1109–1115. doi: 10.1128/IAI.71.3.1109-1115.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrin-Biroulet L, Vignal C, Dessein R, Simonet M, Desreumaux P, et al. NODs in defence: from vulnerable antimicrobial peptides to chronic inflammation. Trends Microbiol. 2006;14:432–438. doi: 10.1016/j.tim.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Voss E, Wehkamp J, Wehkamp K, Stange EF, Schroder JM, et al. NOD2/CARD15 mediates induction of the antimicrobial peptide human beta-defensin-2. J Biol Chem. 2006;281:2005–2011. doi: 10.1074/jbc.M511044200. [DOI] [PubMed] [Google Scholar]
- Uematsu S, Jang MH, Chevrier N, Guo Z, Kumagai Y, et al. Detection of pathogenic intestinal bacteria by Toll-like receptor 5 on intestinal CD11c+ lamina propria cells. Nat Immunol. 2006;7:868–874. doi: 10.1038/ni1362. [DOI] [PubMed] [Google Scholar]
- Boneca IG, Dussurget O, Cabanes D, Nahori MA, Sousa S, et al. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc Natl Acad Sci U S A. 2007;104:997–1002. doi: 10.1073/pnas.0609672104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaput C, Ecobichon C, Cayet N, Girardin SE, Werts C, et al. Role of AmiA in the morphological transition of Helicobacter pylori and in immune escape. PLoS Pathog. 2006;2:e97. doi: 10.1371/journal.ppat.0020097. doi: 10.1371/journal.ppat.0020097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly D, Campbell JI, King TP, Grant G, Jansson EA, et al. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-gamma and RelA. Nat Immunol. 2004;5:104–112. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Keitany G, Li Y, Wang Y, Ball HL, et al. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science. 2006;312:1211–1214. doi: 10.1126/science.1126867. [DOI] [PubMed] [Google Scholar]
- Arbibe L, Kim DW, Batsche E, Pedron T, Mateescu B, et al. An injected bacterial effector targets chromatin access for transcription factor NF-kappaB to alter transcription of host genes involved in immune responses. Nat Immunol. 2007;8:47–56. doi: 10.1038/ni1423. [DOI] [PubMed] [Google Scholar]
- Perrin AJ, Jiang X, Birmingham CL, So NS, Brumell JH. Recognition of bacteria in the cytosol of Mammalian cells by the ubiquitin system. Curr Biol. 2004;14:806–811. doi: 10.1016/j.cub.2004.04.033. [DOI] [PubMed] [Google Scholar]
- Hawn TR, Verbon A, Lettinga KD, Zhao LP, Li SS, et al. A common dominant TLR5 stop codon polymorphism abolishes flagellin signaling and is associated with susceptibility to legionnaires' disease. J Exp Med. 2003;198:1563–1572. doi: 10.1084/jem.20031220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard C, Puel A, Bonnet M, Ku CL, Bustamante J, et al. Pyogenic bacterial infections in humans with IRAK-4 deficiency. Science. 2003;299:2076–2079. doi: 10.1126/science.1081902. [DOI] [PubMed] [Google Scholar]
- Filipe-Santos O, Bustamante J, Haverkamp MH, Vinolo E, Ku CL, et al. X-linked susceptibility to mycobacteria is caused by mutations in NEMO impairing CD40-dependent IL-12 production. J Exp Med. 2006;203:1745–1759. doi: 10.1084/jem.20060085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casrouge A, Zhang SY, Eidenschenk C, Jouanguy E, Puel A, et al. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science. 2006;314:308–312. doi: 10.1126/science.1128346. [DOI] [PubMed] [Google Scholar]
- Khor CC, Chapman SJ, Vannberg FO, Dunne A, Murphy C, et al. A Mal functional variant is associated with protection against invasive pneumococcal disease, bacteremia, malaria and tuberculosis. Nat Genet. 2007;39:523–528. doi: 10.1038/ng1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman HM, Mueller JL, Broide DH, Wanderer AA, Kolodner RD. Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat Genet. 2001;29:301–305. doi: 10.1038/ng756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miceli-Richard C, Lesage S, Rybojad M, Prieur AM, Manouvrier-Hanu S, et al. CARD15 mutations in Blau syndrome. Nat Genet. 2001;29:19–20. doi: 10.1038/ng720. [DOI] [PubMed] [Google Scholar]
- Hawn TR, Wu H, Grossman JM, Hahn BH, Tsao BP, et al. A stop codon polymorphism of Toll-like receptor 5 is associated with resistance to systemic lupus erythematosus. Proc Natl Acad Sci U S A. 2005;102:10593–10597. doi: 10.1073/pnas.0501165102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- Nenci A, Becker C, Wullaert A, Gareus R, van Loo G, et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. 2007;446:557–561. doi: 10.1038/nature05698. [DOI] [PubMed] [Google Scholar]
- Jin Y, Mailloux CM, Gowan K, Riccardi SL, LaBerge G, et al. NALP1 in vitiligo-associated multiple autoimmune disease. N Engl J Med. 2007;356:1216–1225. doi: 10.1056/NEJMoa061592. [DOI] [PubMed] [Google Scholar]
- Bestor TH, Bourc'his D. Genetics and epigenetics of hydatidiform moles. Nat Genet. 2006;38:274–276. doi: 10.1038/ng0306-274. [DOI] [PubMed] [Google Scholar]
- Murdoch S, Djuric U, Mazhar B, Seoud M, Khan R, et al. Mutations in NALP7 cause recurrent hydatidiform moles and reproductive wastage in humans. Nat Genet. 2006;38:300–302. doi: 10.1038/ng1740. [DOI] [PubMed] [Google Scholar]
- Djuric U, El-Maarri O, Lamb B, Kuick R, Seoud M, et al. Familial molar tissues due to mutations in the inflammatory gene, NALP7, have normal postzygotic DNA methylation. Hum Genet. 2006;120:390–395. doi: 10.1007/s00439-006-0192-3. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Wang Y, Hasegawa M, Imamura R, Suda T. PYPAF3, a PYRIN-containing APAF-1-like protein, is a feedback regulator of caspase-1-dependent interleukin-1beta secretion. J Biol Chem. 2005;280:21720–21725. doi: 10.1074/jbc.M410057200. [DOI] [PubMed] [Google Scholar]
- Okada K, Hirota E, Mizutani Y, Fujioka T, Shuin T, et al. Oncogenic role of NALP7 in testicular seminomas. Cancer Sci. 2004;95:949–954. doi: 10.1111/j.1349-7006.2004.tb03182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]